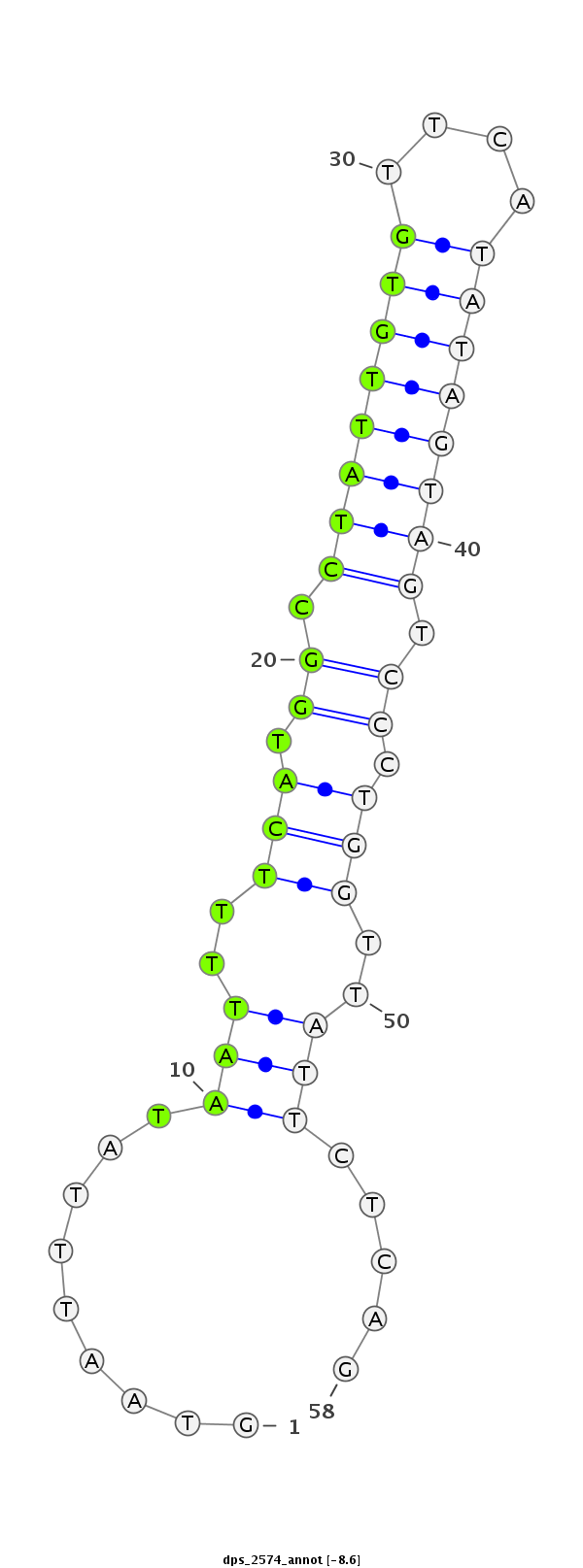
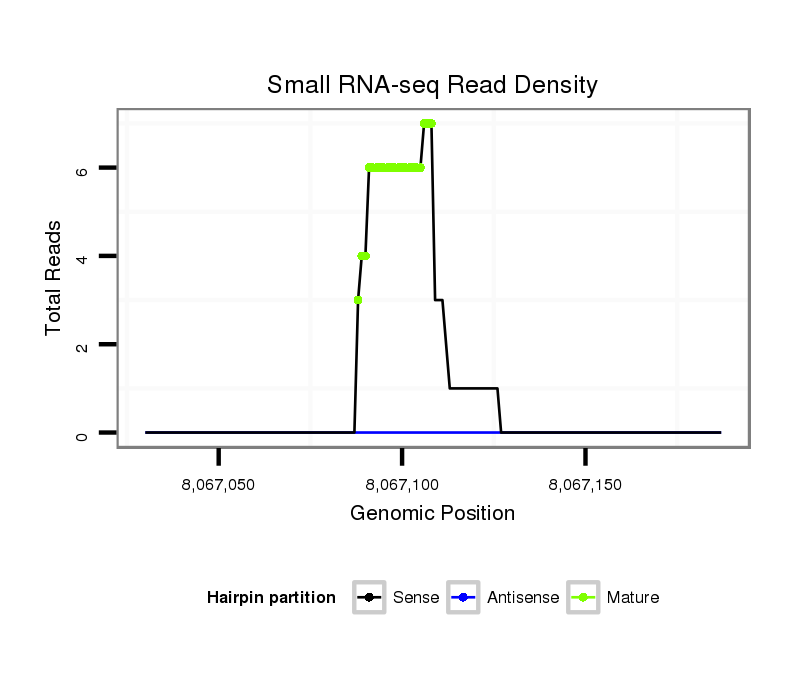
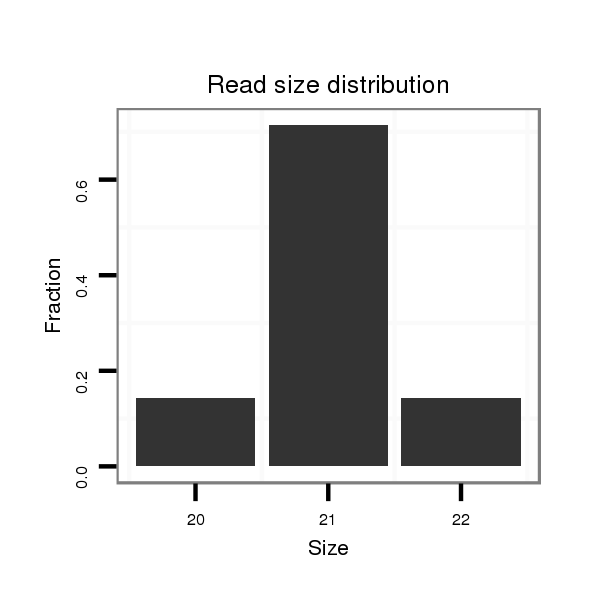
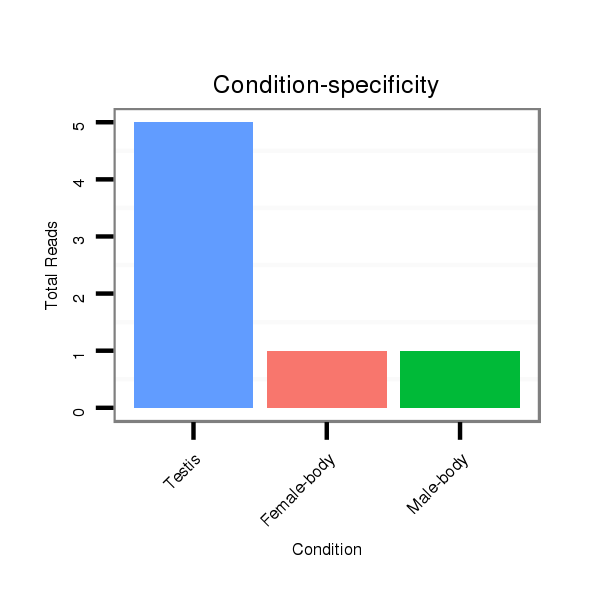
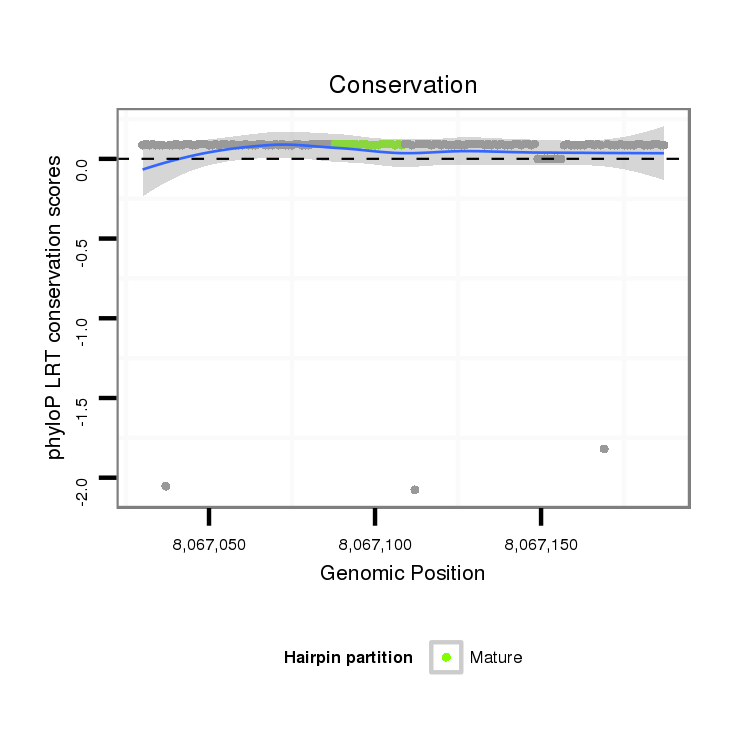
ID:dps_2574 |
Coordinate:XL_group1a:8067080-8067137 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.5 | -8.3 | -8.0 |
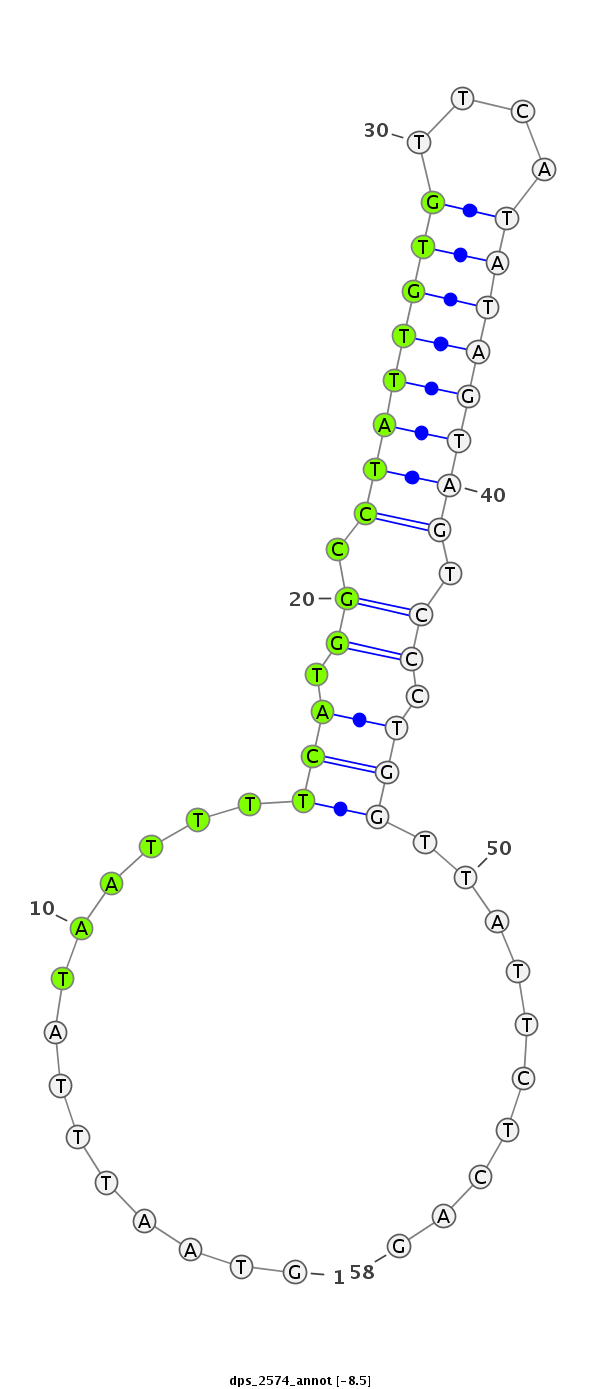 |
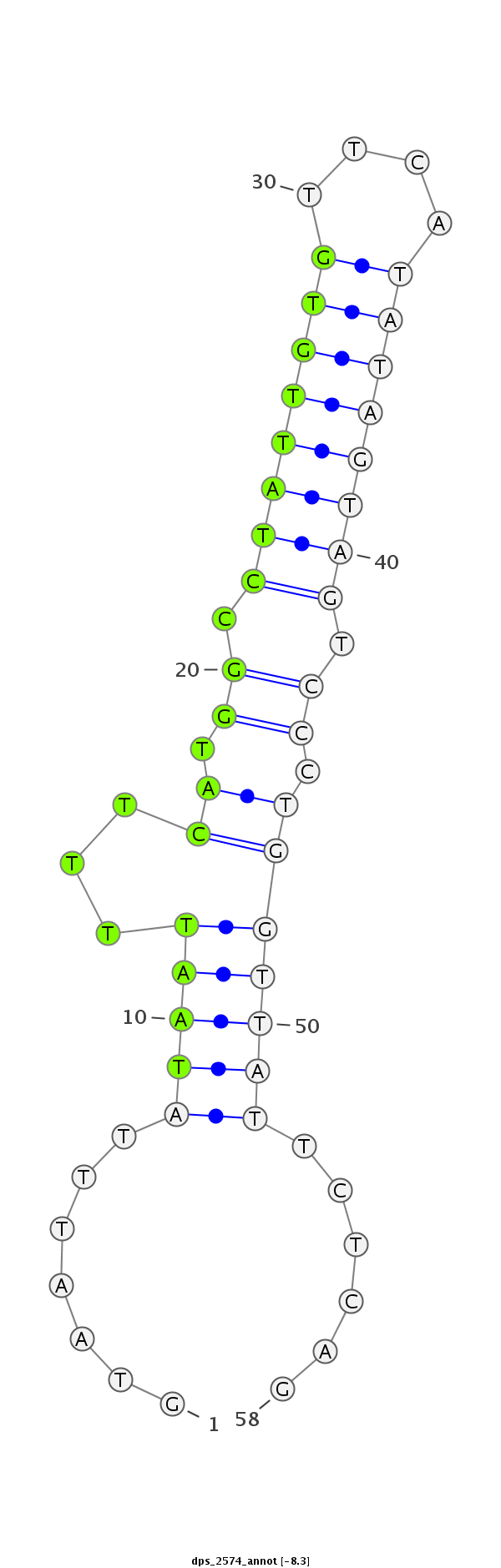 |
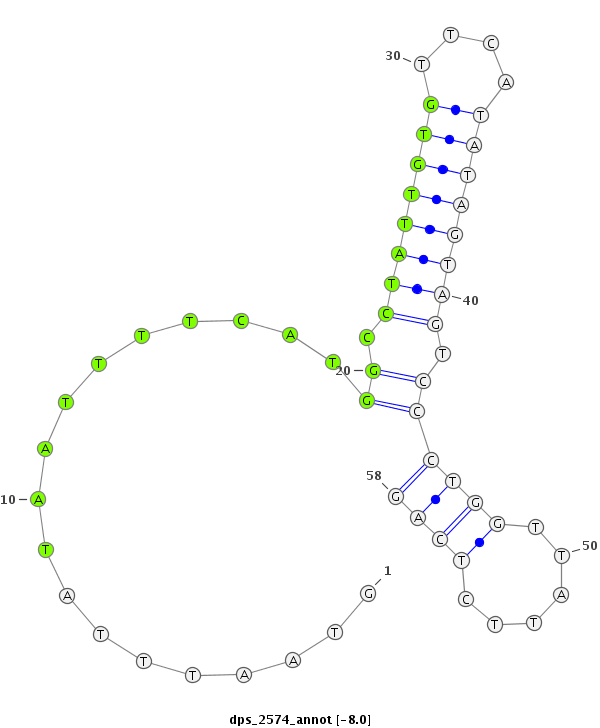 |
CDS [Dpse\GA23081-cds]; exon [dpse_GLEANR_11591:3]; CDS [Dpse\GA23081-cds]; exon [dpse_GLEANR_11591:2]; intron [Dpse\GA23081-in]
No Repeatable elements found
| ##################################################----------------------------------------------------------################################################## GAGAGCGCGGAGGAAGCTAACGAGAAGTGGGAGCGAAGGCTGACGGATCGGTAATTTATAATTTTCATGGCCTATTGTGTTCATATAGTAGTCCCTGGTTATTCTCAGTCGGTACAAGTGCAGCCCTGTCCGGCATCTCAACGGTGGCCATCGCATCG **************************************************.........(((..(((.((.((((((((....)))))))).)).)))..))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
V112 male body |
M040 female body |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................TAATTTTCATGGCCTATTGTG............................................................................... | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ............................................................................GTGTTCATATAGTAGTCCCTG............................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................AATTTTCATGGCCTATTGTG............................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................TATTCATGGCCTATTGTGTTC............................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................TTTTCATGGCCTATTGTGTTC............................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................TTTTCATGGCCTATTGTGTTCA........................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................TCATATAGTAGTCCCTGGTTATCC...................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................GGAGTGGTAGCGAAGTCTG.................................................................................................................... | 19 | 3 | 14 | 0.21 | 3 | 0 | 0 | 3 | 0 |
| .................................CGAAGGCTGGCGGATCGTAA......................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |
| .......................GACGTGGTTGCGAAGGCT..................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
CTCTCGCGCCTCCTTCGATTGCTCTTCACCCTCGCTTCCGACTGCCTAGCCATTAAATATTAAAAGTACCGGATAACACAAGTATATCATCAGGGACCAATAAGAGTCAGCCATGTTCACGTCGGGACAGGCCGTAGAGTTGCCACCGGTAGCGTAGC
**************************************************.........(((..(((.((.((((((((....)))))))).)).)))..))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343916 embryo |
|---|---|---|---|---|---|---|
| ........................TTCACACTCGTTTCCGAGTG.................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1a:8067030-8067187 + | dps_2574 | GAGAGCGCGGAGGAAGCTAACGAGAAGTGGGAGCGAAGGCTGACGGATCGGTAATTTATAATTTTCATGGCCTATTGTGTTCATATAGTAGTCCCTGGTTATTCTCAGTCGGTACAAGTGCAGCCCTGTCCGGCATCTCAACGGTGGCCATCGCATCG |
| droPer2 | scaffold_15:425077-425226 + | GAGAGCGAGGAGGAAGCTAACGAGAAGTGGGAGCGAAGGCTGACGGATCGGTAATTTATAATTTTCATGGCCTATTGTGTTCCTATAGTAGTCCCTGGTTATTCTCAGTCGGTACAAGT--------GTCCGGCATCTCTACGGTGGCCATCGCATCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 04:55 AM