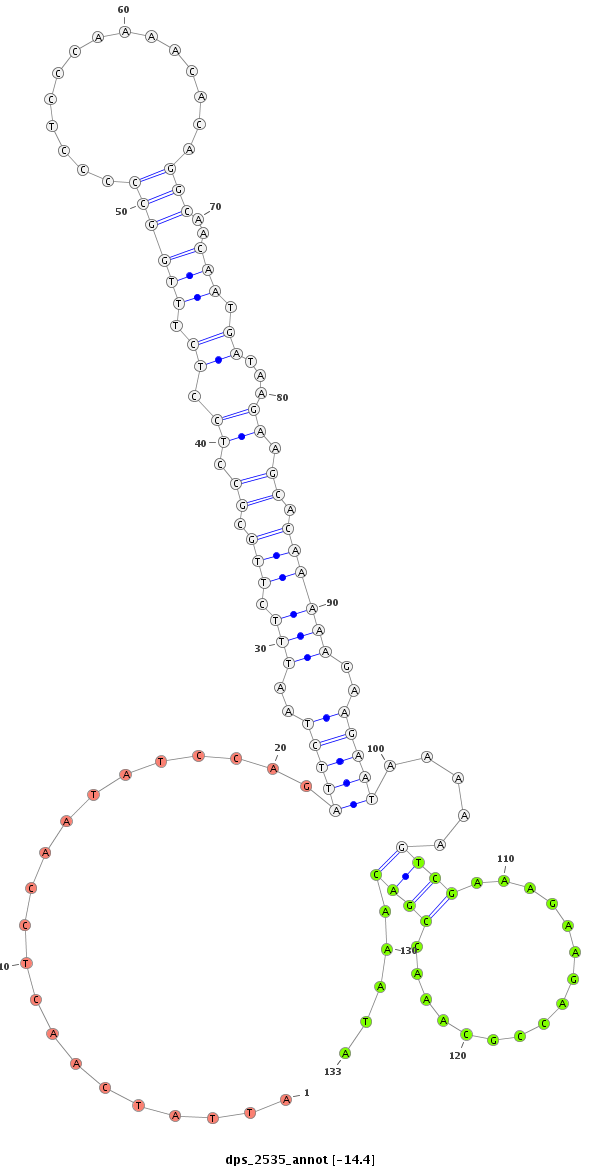
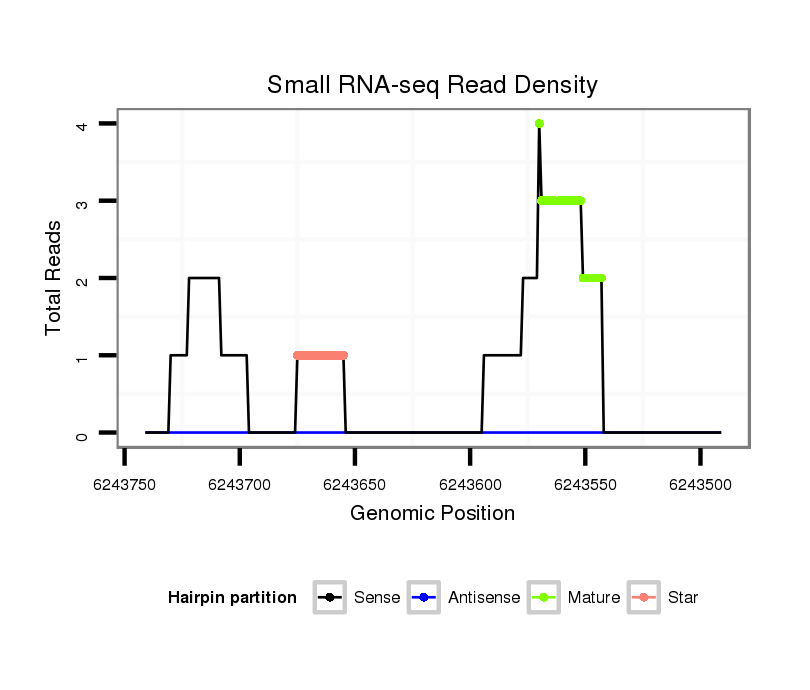
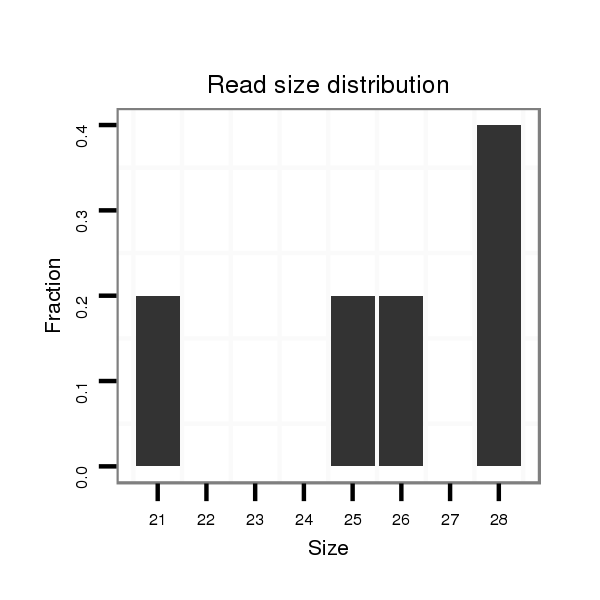
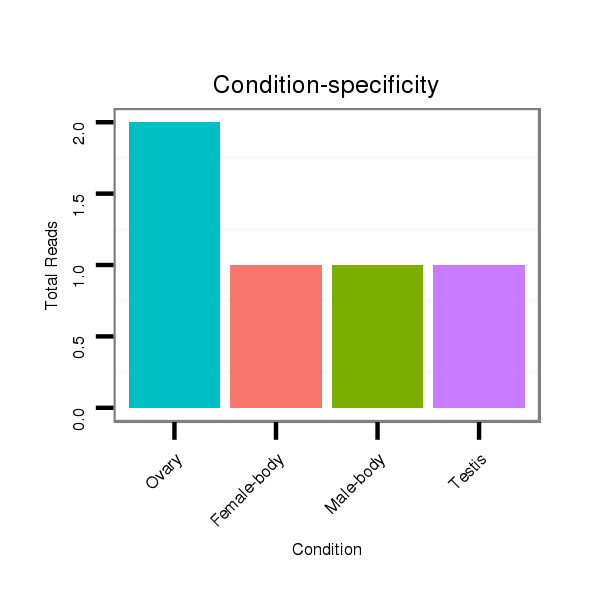
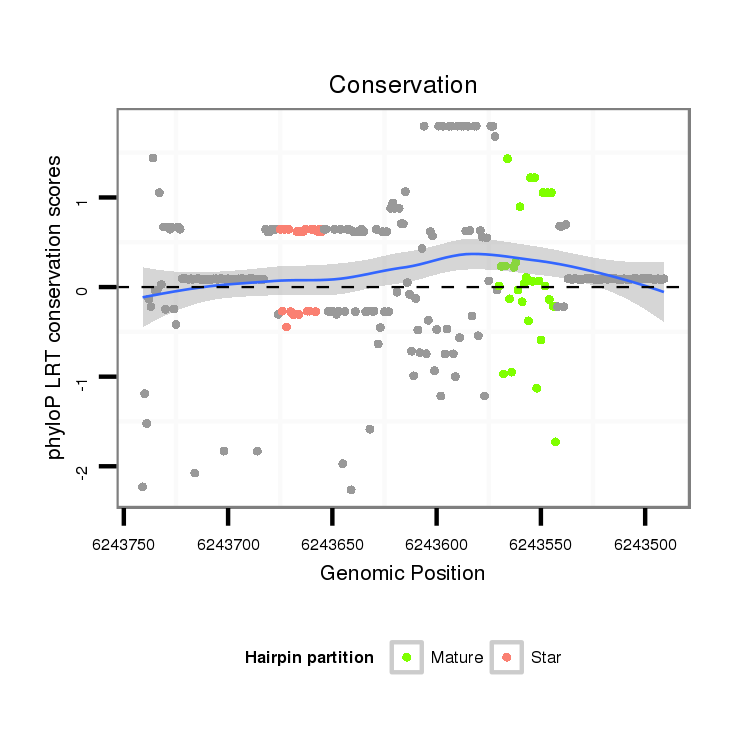
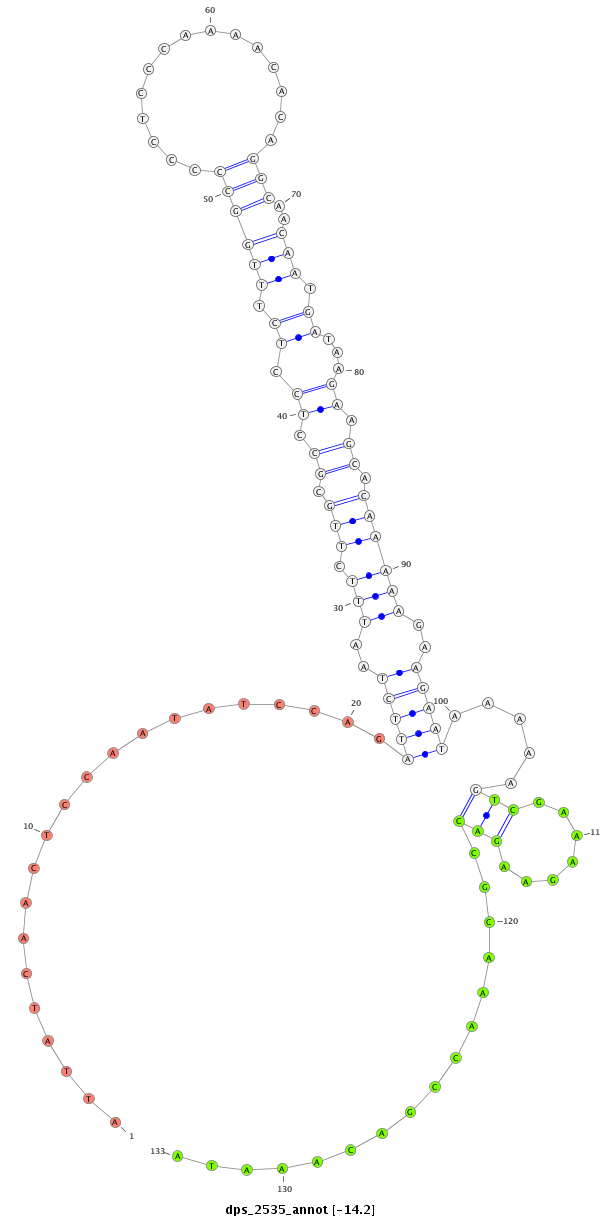
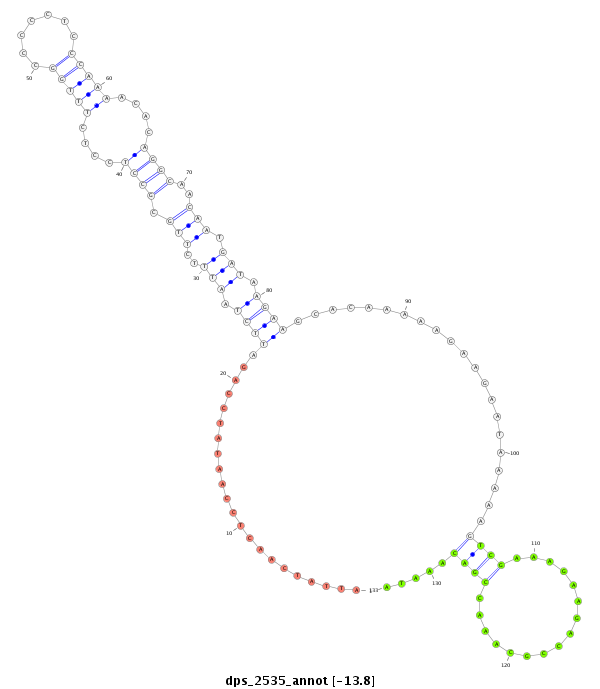
ID:dps_2535 |
Coordinate:XL_group1a:6243541-6243691 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -14.2 | -13.8 | -13.7 |
 |
 |
 |
exon [dpse_GLEANR_10813:1]; CDS [Dpse\GA15320-cds]; intron [Dpse\GA15320-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAGCCCAAACAGTGAATACAATGTGAATGGCCCCTTGAAGCTGGCCAAAGGTAAGGGGCTGGAAATATTATCAACTCCAATATCCAGATTCTAATTTCTTGCGCCTCCTCTTTGGCCCCCTCCCAAAACACAGGCAACAATGATAAGAAGCACAAAAAGAAGAATAAAAAGTCGAAAGAAGACCGCAAACCGACAAATACCGGATTGAAGGGATATTCCGGAACAAATCCCTCCCTATCTTATATGCAGCA ******************************************************************.....................(((((..(((.(((.((.((.((.((((((...............)))..))).))...)).)).))))))..))))).....((((................)))).....**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
SRR902010 ovaries |
V112 male body |
SRR902011 testis |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................TCGAAAGAAGACCGCAAACCGACAAATA.................................................... | 28 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...................AATGTGAATGGCCCCTTGAAGCTGGC.............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AAGCACAAAAAGAAGAATAAAAAGT............................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........GTGAATACAATGTGAATGGCCC.......................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....CAAACAGTGAATACAATGTGACTGGC............................................................................................................................................................................................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TAAAAAGTCGAAAGAAGACCGCAAAC............................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................ATTATCAACTCCAATATCCAG.................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TTCTTGCGCCTCCTTTTGGGCCC..................................................................................................................................... | 23 | 2 | 4 | 0.50 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................TTCTTGCGCCTCCTTTTTGGCC...................................................................................................................................... | 22 | 1 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ............................................CCGAAGGTGCGGGGCTGGAAA.......................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................TCTTGCGCCTCCTTTTGGGCCC..................................................................................................................................... | 22 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ATAAGGAGCGCAAAAAGAAGG........................................................................................ | 21 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................ATACCGGCTGGAAGAGATA.................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AAATACCGGATTGAAGATG...................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GTCGGGTTTGTCACTTATGTTACACTTACCGGGGAACTTCGACCGGTTTCCATTCCCCGACCTTTATAATAGTTGAGGTTATAGGTCTAAGATTAAAGAACGCGGAGGAGAAACCGGGGGAGGGTTTTGTGTCCGTTGTTACTATTCTTCGTGTTTTTCTTCTTATTTTTCAGCTTTCTTCTGGCGTTTGGCTGTTTATGGCCTAACTTCCCTATAAGGCCTTGTTTAGGGAGGGATAGAATATACGTCGT
****************************************************.....................(((((..(((.(((.((.((.((.((((((...............)))..))).))...)).)).))))))..))))).....((((................)))).....****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................TTAGGGAGAGATAGTATAT....... | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| .................................................................................................................................................TCTTCGTGTTCTTCTTCTTA...................................................................................... | 20 | 1 | 4 | 0.25 | 1 | 1 | 0 |
| ..................................................................................AGGTTTAAGCTTCAAGAACG..................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 1 | 0 |
| .........................................................................................AGATTATAGAGGGCGGAGG............................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1a:6243491-6243741 - | dps_2535 | CAGC---CCAAACAGTGAATACAATGTGAATGGCCCCTTGAAGCTGGCCAAAGGTAAGGGGCTGGAAATATTATCAACTCCAATATCCAGATTCTAATTTCTTGCGCCTCCTCTTTGGCCCCCTCCCAAAAC-ACAGGCAACAATGATAAGAAGCACAAAAAGAAGAATAAAAAGTCGAAAGAAGACCGCAAACCGACAAATACCGGATTGAAGGGATATTCCGGAACAAATCCCTCCCTATCTTATATGCAGCA |
| droPer2 | scaffold_25:1084909-1085159 + | CAGC---CCAAACAGTGAATACAATGTGCATGGCCCCTTGAAACTGGCCAAAGGTAAGAGGCTGGAAATATTATCAACTCCAATATCCAGATTCTAATTGCTTTCGCCTCCTTTTTGGCCCCCTCCCAAAAC-ACAGGCAACAATGATAAGAAGCACAAAAAGAAGAATAAAAAGTCGAAAGAAGACCGCAAACCGACAAATGCCGGATTGAAGGGATATTCCGGAACAAATCCCTCCCTATCTTATATGCAGCA | |
| droWil2 | scf2_1100000004585:6105907-6105951 - | ACAC---AC--------------------------------------------------------------------------------------------------------------------------------ACACAAAACAAAGGAAGCAAAAAAAAAAAAACAAAAAGT------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12875:16285231-16285277 - | CCAA-----------------------------------------------------------------------------------------------------------------------------ATT-GCAGGCAACTACAAAAAAAAACAAAAAAAAAAAAACAAAA---------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6473:3593945-3594010 + | TTGACGCTCAAAT---------------------------------------------------------------------------------------------------------------------AA-AACAGCAACAATAATACAAACAACAAAATGAAAGAAATAAAGAAAAAAGAA----------------------------------------------------------------------- | |
| droGri2 | scaffold_15245:264527-264596 + | AAAG-------------------------------------------------------------------------------------------------------------------------------C-AACAACAGCAACAACAACAACAACAAGAAGAAGAAGAAAAAAACCAAGACA---CGCAAAGCTATAAA------------------------------------------------------ | |
| droBip1 | scf7180000395934:16621-16639 - | CAGC---TCGGACAATGAGGAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302300:102391-102458 - | GGC-----------------------------------------------------------TGGAAAAACTCTTTTCACCAGCATTCAGATCTCATCTTTTTCCACCTCTCCCCTCTCTCTCTCTC-------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000414375:366577-366582 + | TAGC---TC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEug1 | scf7180000409553:220162-220239 + | CCCC----------------------------------------------------------------------------------------------------------------------CTCTCAAAACACAAAACAAAATTTACAAAAAAAAAAAAAAGAAAAAAAGAAAAACAAAGAATAATAATAAACCG----------------------------------------------------------- | |
| droSec2 | scaffold_34:291814-291893 + | AAAA-------------------------------------------------------------------------------------------------------------------------------C-TCAGCGACGAAAAATACGAAATACAAAAAAAATATAGAAAAAACGCAGCAAAACAGAAGAGAAACACACATCGAA----------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/18/2015 at 04:51 AM