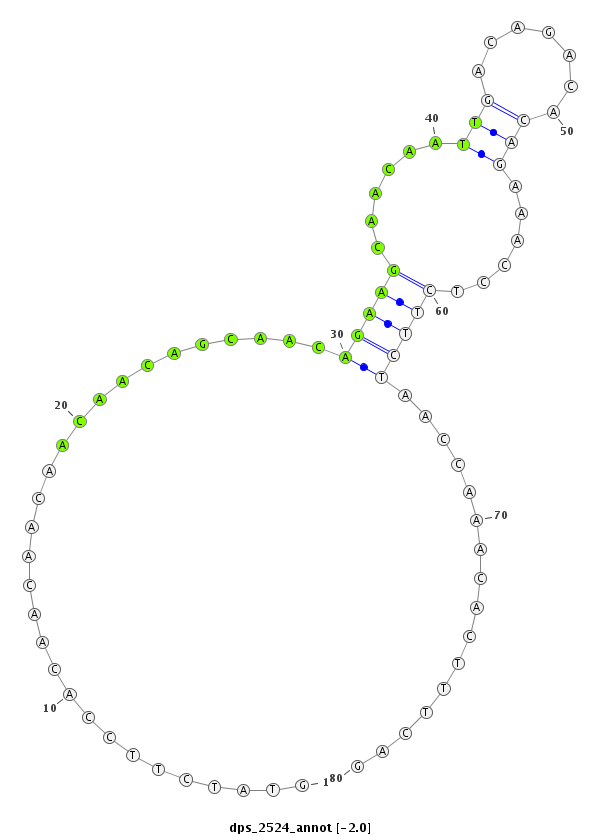
| GCAGGAACAGCAACAACAGCAGCAGCAACAACAACAACAG--C--------------A---------------GC---AGCAACAATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGCAGCAG---GAACAGCA---AC--A-AC---AGCAGCAG------------------CAA---CA------------------ATATCA--------ACA---------G------CAACAGCAACAACAGCAGCAGCAACAGCAA-CAACAGCAGCAGCAGCAGCAAC | Size | Hit Count | Total Norm | Total | M020
Head | M045
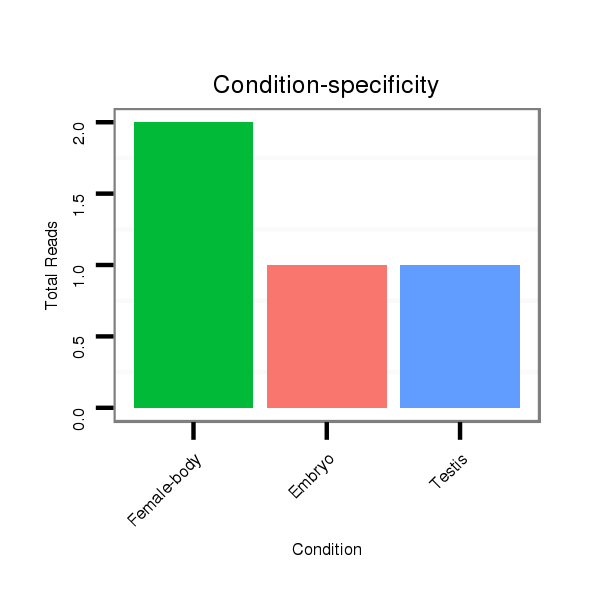
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ..........................................................................................................................................................C--------A--------ACAGCAGCAG---GAACAGCA---AC--A-AC---.......................................................................................................................................... | 25 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAA......................................... | 23 | 20 | 0.60 | 12 | 0 | 12 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------A............................................................................................................................................................................ | 23 | 20 | 0.60 | 12 | 0 | 12 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------............................................................................................................................................................................. | 22 | 20 | 0.45 | 9 | 0 | 9 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACA.......................................... | 22 | 20 | 0.45 | 9 | 0 | 9 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAAC........................................ | 24 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGC........................................................................................................................................................................ | 27 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAACAGC..................................... | 27 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ..........................AACAACAACAACAG--C--------------A---------------GC---A.......................................................................................................................................................................................................................................................................... | 19 | 20 | 0.25 | 5 | 0 | 0 | 0 | 5 | 0 |
| ........................................................................................................................................................AAC--------A--------ACAGCAGCAG---GAACAGCA---AC--A-............................................................................................................................................... | 25 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------AC........................................................................................................................................................................... | 24 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAACAG...................................... | 26 | 20 | 0.20 | 4 | 0 | 3 | 0 | 1 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAG......................................................................................................................................................................... | 26 | 20 | 0.20 | 4 | 0 | 3 | 0 | 1 | 0 |
| .....................................................................................................................................................AGCAAC--------A--------ACAGCAGCAG---GAACAGCA---..................................................................................................................................................... | 25 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................CAACAACAACAG--C--------------A---------------GC---AGCAACAAT.................................................................................................................................................................................................................................................................. | 25 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................A---------G------CAACAGCAACAACAGCAGCAG................................ | 23 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................AGCAACAGCAA-CAACAGCAGCAG.......... | 23 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ..................................................................................................................AGC------------A-----------------ACAGCAAC--------A--------ACAGCAGCAG---................................................................................................................................................................ | 23 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAACA....................................... | 25 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACA.......................................................................................................................................................................... | 25 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| .............................AACAACAACAG--C--------------A---------------GC---AGCAAC..................................................................................................................................................................................................................................................................... | 21 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ...................................................................................................................................................ACAGCAAC--------A--------ACAGCAGCAG---G............................................................................................................................................................... | 20 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGCAG...................................................................................................................................................................... | 29 | 19 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAACAGCAG................................... | 29 | 19 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......CAGCAACAACAGCAGCAGC............................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ...............................................................................................................................................................................................................................................................................................................ACAGCAGCAGCAACAGCA........................ | 18 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .....................GCAGCAACAACAACAACAG--C--------------................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAACAGCA.................................... | 28 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................AA---CA------------------ATATCA--------ACA---------G------CAACAGCAACA.......................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAAC........................................... | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ATCA--------ACA---------G------CAACAGCAACAACAG...................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................................................CAGCA---AC--A-AC---AGCAGCAG------------------C............................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .........................CAACAACAACAACAG--C--------------A---------------GC---AGC........................................................................................................................................................................................................................................................................ | 22 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| .................................................................................AACAATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------............................................................................................................................................................................. | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................ATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGCA....................................................................................................................................................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGCA....................................................................................................................................................................... | 28 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................ATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAG......................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................CAGCAA-CAACAGCAGCAGC......... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .....................................................................................................................................................................................................................................................................ATCA--------ACA---------G------CAACAGCAACAACAGCA.................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AGCAGCAG---GAACAGCA---AC--A-............................................................................................................................................... | 19 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................AACAG--C--------------A---------------GC---AGCAACAATATC---------------AAC------AG..................................................................................................................................................................................................................................... | 26 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................A-AC---AGCAGCAG------------------CAA---CA------------------ATATCA--------ACA---------G------..................................................... | 26 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................ACAG--C--------------A---------------GC---AGCAACAATATC---------------AAC------....................................................................................................................................................................................................................................... | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AC---AGCAGCAG------------------CAA---CA------------------ATATCA--------AC...................................................................... | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................AGCAAC--------A--------ACAGCAGCAG---G............................................................................................................................................................... | 18 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................C--------------A---------------GC---AGCAACAATATC---------------AAC------AGC------------A-----------------A..................................................................................................................................................................................................... | 24 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ATCA--------ACA---------G------CAACAGCAACAACAGCAGCAG................................ | 29 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................CAGCAG------------------CAA---CA------------------ATATCA--------ACA---------G------CAA.................................................. | 24 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................ATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGCAGCAG---................................................................................................................................................................ | 29 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................A---------------GC---AGCAACAATATC---------------AAC------AG..................................................................................................................................................................................................................................... | 20 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGCAG...................................................................................................................................................................... | 27 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................TATCA--------ACA---------G------CAACAGCAACAACAGCAG................................... | 27 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................AGCAG------------------CAA---CA------------------ATATCA--------ACA---------G------..................................................... | 20 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....AACAGCAACAACAGCAGCAGCA.............................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............AACAGCAGCAGCAACAACAAC...................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................ACAGCA---AC--A-AC---AGCAGCA................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................GCAA-CAACAGCAGCAGCAGCAGC... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................CAGCAACAACAACAACAG--C--------------A---------------GC---A.......................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................CAACAACAACAACAG--C--------------A---------------GC---AGCAA...................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................C------------A-----------------ACAGCAAC--------A--------ACAGCAGCAG---................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................ATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACA.......................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................A--------ACA---------G------CAACAGCAACAACAGCA.................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................................................................................................................AGCAGCAGCAACAGCAA-CAACAGC............... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................CAACAACAGCAGCAGCAACAG.......................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................CAGCAGCAACAGCAA-CAACAG................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................CAGCAA-CAACAGCAGCAGCAGCAGCA.. | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................AGC------------A-----------------ACAGCAAC--------A--------ACAGCAGCA.................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................CAACAGCAA-CAACAGCAGCAGCAGCAG.... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................GCAGCAACAACAACAACAG--C--------------A---------------GC---........................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................A---------G------CAACAGCAACAACAGCAGCAGCAAA............................ | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................AACAACAACAG--C--------------A---------------GC---AGCA....................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................GCAGCAGCAACAACAACAACAG--C--------------................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................AGCAA-CAACAGCAGCAGCAGC...... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................................................ATATCA--------ACA---------G------CAACAGCAACAAC........................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................CAACAGCAGCAGCAACAGC......................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................CAG------------------CAA---CA------------------ATATCA--------ACA---------G------CAACAGC.............................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................AACAATATC---------------AAC------AGC------------A-----------------ACAGCAA............................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................TATCA--------ACA---------G------CAACAGCAACAACAGC..................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................CAACAGCAACAACAGCAGCAG................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................AACAGCAA-CAACAGCAGCAGCA........ | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................ATATCA--------ACA---------G------CAACAGCAACAACAGC..................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............CAACAGCAGCAGCAACAACAACAAC................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACA.......................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................CAACAACAACAG--C--------------A---------------GC---AGCAACAA................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................AGCAACAGCAA-CAACAGCAGC............ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................AACAATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------A............................................................................................................................................................................ | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................CA--------ACA---------G------CAACAGCAACAACAGCAGCAGCA.............................. | 29 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................CAACAATATC---------------AAC------AGC------------A-----------------ACAGCAA............................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................AACAACAGCAGCAGCAACAGCA........................ | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................ACAGCAGCAGCAACAGCAA-C..................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAACAGTT.................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................A------------------ATATCA--------ACA---------G------CAACAGCAACAT......................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................CA------------------ATATCA--------ACA---------G------CAACAGCAACAAC........................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................AA---CA------------------ATATCA--------ACA---------G------CAACAGCAA............................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................A---CA------------------ATATCA--------ACA---------G------CAACAGCAACA.......................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................AGCAA-CAACAGCAGCAGCAGCAGCA.. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................CAACAGCAA-CAACAGCAGC............ | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................AA---CA------------------ATATCA--------ACA---------G------CAACAGCAACAA......................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AGCAGCAACAGCAA-CAACAGCAGC............ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................AGCAA-CAACAGCAGCAGCAG....... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................CAA-CAACAGCAGCAGCAGCA..... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................AACAGCA---AC--A-AC---AGCAGCAG------------------CA.............................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................GCAACAACAACAACAG--C--------------A---------------GC---AGC........................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................GCAA-CAACAGCAGCAGCAGCA..... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................AGCAACAACAGCAGCAGCAACAGC......................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................AA---CA------------------ATATCA--------ACA---------G------CAACAGCAAC........................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................AATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------T............................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................ACAGCAA-CAACAGCAGCAGCAGCA..... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................AGCAACAACAACAACAG--C--------------A---------------GC---AGCAAC..................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................................................................................................GCAGCAACAGCAA-CAACAGCAGCAGC......... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................CAACAGCAACAACAGCAGC.................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............AACAGCAGCAGCAACAAC......................................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGC........................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................TATCA--------ACA---------G------CAACAGCAACAACA....................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................CAA---CA------------------ATATCA--------ACA---------G------CAACAGCAA............................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................ATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------AC........................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................ATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------ACAGC........................................................................................................................................................................ | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................A---------G------CAACAGCAACAACAGCAGCAGC............................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................AA---CA------------------ATATCA--------ACA---------G------CAACAGCA............................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ATCA--------ACA---------G------CAACAGCAACAACA....................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................AGCAACAGCAA-CAACAGCAGCAGC......... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................ACAATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------A--------............................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................AACAATATC---------------AAC------AGC------------A-----------------ACAGCAAC--------...................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................CAGCAACAACAGCAGCAGCAACA........................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |