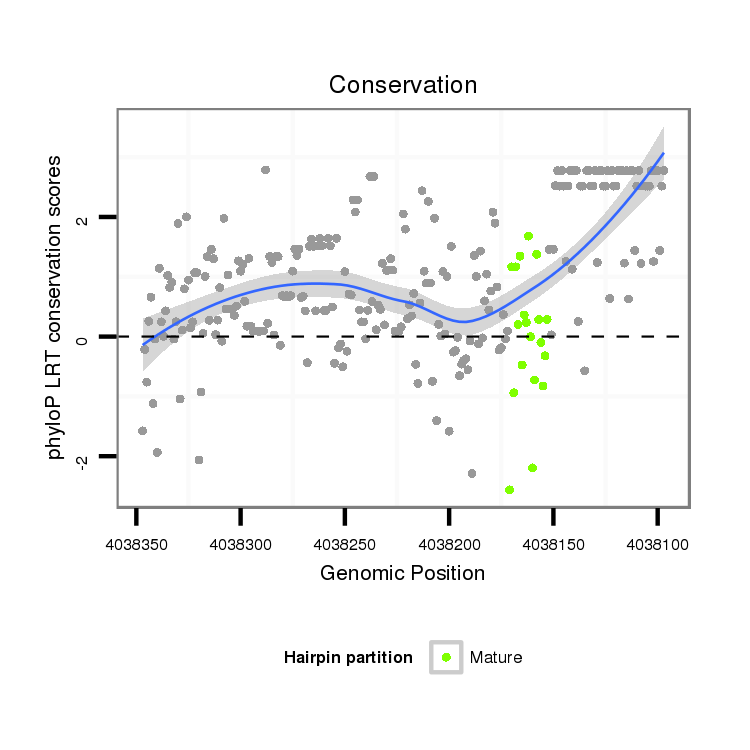
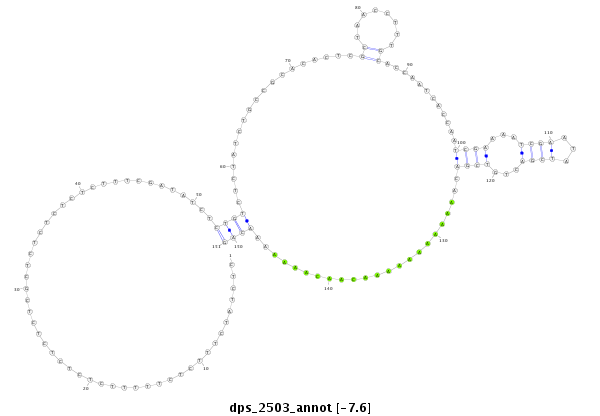
| dp5 |
XL_group1a:4038097-4038347 - |
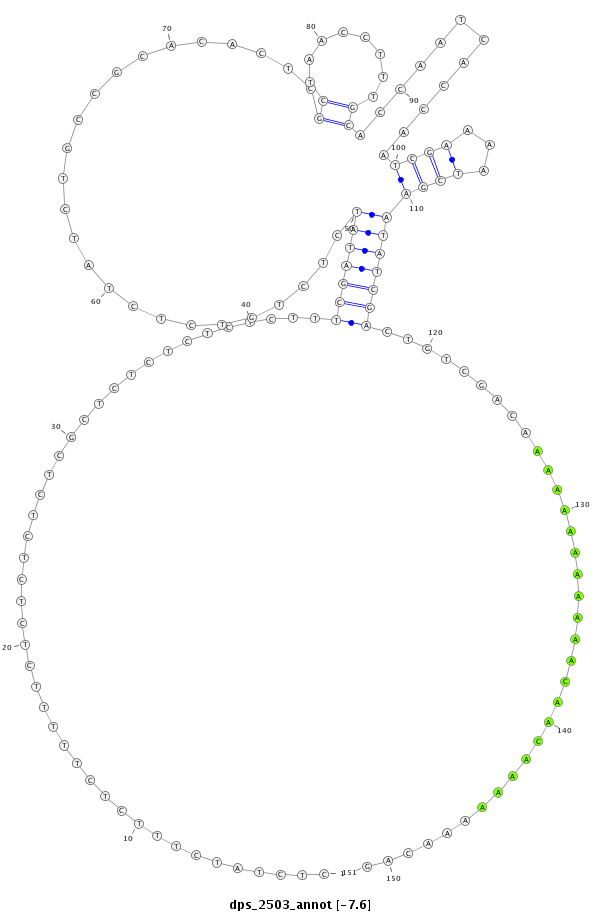
dps_2503 |
CTGTACAG-------AAGATAAT---G--TAATTTTA------------------------------------TTAAAT-------TT--------------------------------------------------TTT-------------------------------------------TTC--TTTTT-----GCTCTTCTCTATCTTTCTCTTTTTCTCTCTCTCTCGCTCTCTC--TCTCTTTCGAT----ATCTCT--------G-----------------------------------TCTCTATC----TG------CCGCACACTCGCT---AACC---------------------------------------------TTT----------------------------------------------------------GCACCAATCACC---------A------------------------------------------------------------------------------------ATCGAAAATCGA----AT-AT------------------------------CGACTGT---CG----A--------C-----------AAAAAAAAAAAA--------------------------------------------------------CAAC-------AAAAAAACAGTCGCAGTTCCGCTTCGATAACGAATGCTATCTGAATCTGCCGGACAATCA |
| droPer2 |
scaffold_11:605149-605396 - |
|
CTGTACAG-------AAGATAAT---G--TAATTTGA------------------------------------TTCAAT-------TT--------------------------------------------------TTT-------------------------------------------TTC--TTTTT-----GCTCTTCTCTATCTTTCTCTTTTTCTCTCTCT--------CTC--TCTCTTTCGAT----ATCTCT--------G-----------------------------------TCTCTATC----TG------CCGCACACTCGCT---AACC---------------------------------------------TTT----------------------------------------------------------GCACCAATCACC---------A------------------------------------------------------------------------------------ATCGAATATCGA----AT-AT------------------------------CGACTGT---CG----A--------C------TGTCGACAAAAAAAAAA--------------------------------------------------------CAAC-------AAAAAAACAGTCGCAGTTCCGCTTCGATAACGAATGCTATCTGAATCTGCCGGACAATCA |
| droWil2 |
scf2_1100000004401:483995-484284 - |
|
AAGAAAAAAAAAGAAAA-TTAATGTAATG---TTTGTT-------------------------------------TTATCTTCTACTTTCTTTTTCGTCTTTTT----------------------------CA-------------------CACTCTCTTTGT-----------------------------------CTCTTCTC------T------------CTCT--CTCTCGCTCTCGCTCTATTA--------------TCCTGTGTGTTT--C--GCT--------------------GCGTCTTGTG----TA------C--------------TTCAA----TTTGCACCTGCAACAAAATCAAATCAAAATGCAAATCTAAACTT-------------------------------------------------------AATGCACA-------TGCTAAACCA-------AAACAAAAAAAA------AAAAA------------------------------------C-------------------------------------------------------------CAAC--CACA-------------------------------------------------------------------------------------------------------------------CTGAACAGTCGCAATTTCGTTTCGATAACGAATGCTATCTGAATCTGCCGGATAATCA |
| droVir3 |
scaffold_12928:3152629-3152979 + |
dvi_8702 |
TTATAACA-------ATAATTAT---G--TG-----------------------------------TATTTTTTTCTAT-------TTTTTTCTTGTTCACTTG----------------------------TG-------TGCGCTCTCTCTCTCTCTCTCTGTCTCTTA----------------------------------CTT------T------------CTCT--TACTCTCTCTCTCTCTTTTT------------------CTCTTTTTG-----------------------------TCT---------CA-ATGCGCTGCAC---C-----A--AC---------------------------------------------TTTGCACCAATGCCACCAACGAC-----------------------------------------------------------A-ACAACA-------ACAAC-AACAA-----CAAC-------------------------------------------------AACAACACT-------------------------------------AAC--AACGCTGT---GTTTGCGTCTGTCTGTTGTTAC----------GACCCACACATCCGCACACACACATGCGCGCACACATGCGCACACACACGCGCACACACACGCT--C------GCACAAAACAGTCGCAATTCCGTTTCGATAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droMoj3 |
scaffold_6473:384935-385174 - |
|
ACA----A-------TA-ATTAC---ATGTATTTTTT------------------------------------TTCC------AATTCTTTTTTCTTTTTCTTT----------------------------TG-------------------TATTTTACTTGT-------------------------CCCC-----GCTCTCTC-------T------------------CTCTCTCTC--TCTCTTTCTCT----TTCTCT--------C----------CTT--------------------CTCTCTTGTTCTCGTG------CCGCAC---C-----A--AT---------------------------------------------TTT----------------------------------------------------------G--------------------------------------------------------------------------TGTCCGTCATACACACACACACAC---------------ATCAACAC-CAAT------------------------------AC-------------------------------------------ACAC--------------------------------------------------------ACA-------CACACACACAGTCGCAATTCCGCTTCGATAACGAGTGCTATCTGAATCTTCCGGACAATCA |
| droGri2 |
scaffold_14853:2274120-2274451 + |
|
ATGTATTT--------------------------TTG------------------------------------TTAAC-----TATTTTCTTTTTTTTTTCCTG----------------------------TG-------C----------------------------GCTCTTTCTT----------------------------------TCTCTTTCTCTCTCTCTCTCTCTCTCTC--TCTCTCTTT------------TTCTCTTTGTCTCTTTC---TC--------------------TCTTTTTGTC----TAAATGCGCTGCAC---TG-----A-AC---------------------------------------------ATTGCACCATC------AAACAA-CAACAA---------------------------------------------------------------------------CGA-----CAATAACAATAATAACAACGATGTGTC-----------------------------------------------------------------------------------------T--------GTATGTTGTTCCTTT---AACAAACACACACACACACACACACACCTGCAATGACA---ACTCACACACACACACACACACACGCA-------TTCACGCATACAGTCGCAATTCCGTTTCGATAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droAna3 |
scaffold_13117:2550255-2550522 + |
|
TTTTTTAA-------AAGT------------------------------ATATACATA--AA------CATAT------------------------------------------------------------ATATATTT-------------------------------------------TTC--TTTTT-----GCTCTTTTT------TCTCCTTT----------------------------------------TCTCTCTCCGTGTGTTC-----AAAAATAATAAA-AAATACTACTACTTCCTGTCCT-ACG------CTGCAC---CGCTAACAAAA-------------------------AACCAAAATACGAATCAAAAATT----------------------------------------------------------GCACTACA----C-----TTTA-------AAA--------AAA----A-A--------------------------------------C---------------------------------------------------------------AC--ATCACTGAACCAAATGAA---------------A------------------------------------------------------------------ATATGTT-------TAAACCAAATCAGTCGCAGTTCCGATTCGACAACGAGTGCTATTTGAACCTGCCGGACAACCA |
| droBip1 |
scf7180000396696:575538-575838 - |
|
AAAA---G-------AT-AAATT---G--TGAATTGTTTTTTAAAAG-TATATACATGAACATACATATATAT------------------------------------------------------------ATATATTT-------------------------------------------TTC--TTTTT-----GCTCTTTTT------TCTCCTTT----------------------------------------TCTCTCTCCGTGTGTTC--C--AAAAATATTAAAAAAATACTACTACTTCCTGTCCT-ACG------CTGCAC---CGCTAAC---C-------------------AAAAAAAACCAAAATACGAACCAAAAATT----------------------------------------------------------GCACCACA----C-----TTTG-------AAA--------AAA----AAA--------------------------------------CA-------------------------------------------------------------CAC--ATCACTGT---GAATGAA---------------A------------------------------------------------------------------ATATCTT-------TAAACCAAATCAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTGAACCTGCCGGACAACCA |
| droKik1 |
scf7180000302698:1330795-1331089 + |
|
CTGTACAG-------AA-ATAAT---G--TGATTTGTT------------------------------TA--------------------------------TACGTATTATACATATTTTATATATATATATATATAT--T----------------------------GCCGTTTCTCTT--TTC--TTTTT-----GCTCTTTTT------T----------------------------------------CTC----TCT-----------------------------------------------CTGTC----CG------CTGCAC---CGCTAAAAAAAATAAATACTACCAACAAAAAAATTAAA---------------------------------------------------------------------A--CAAAAATGCACCAAAAATC---------CAACACCAAAA--------AAA----A-A--------------------------------------CACAAAAACAC----------------------------CATAAAAAAATTATATAAAAAAACAAC--AAC---------------------------------------------AA--------------------------------------------------------CAAC-------AAAAAAACAGTCGCAGTTTCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droFic1 |
scf7180000454045:225024-225272 + |
|
CTGTACAG-------A--ATAAT---G--TCATTTGTT------------------------------CA--------------------------------TA----------------------------TA-------T----------------------------GCCGTTTCTCTTTTTTC--TTTTC-CTTTGCTCTTTTT------T----------------------------------------CTC----TCTCTTTCCGTGTTTTTTC---TCA--------------------ACTTCCTGTC----CG------CCGCAC---TGCTACAAAAAA-------------------------AT----CTACTA--CAA--AAT----------------------------------------------------------GCACCATA----C-----AC-A-ACAACA-------AAAAT------AAAAAC--C-------------------------------------------------AACCACATTTCA---------------------------------------AACGCTGT---GT-------------------------------------------------------------------------------------------------------AACAAAAACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droEle1 |
scf7180000491272:332116-332346 - |
|
CTGTACAG-------AA-ATAAC---G--TCGTTTGTT------------------------------CA--------------------------------TA----------------------------TA-------T----------------------------GCCGTTTCTCTT--TTC-TTTTTT-----GCTCTTTTT------T----------------------------------------CTCTC--TCTCTTTTCGTGTTTTT--TTCAAA--------------------ACTTCTTGTC----CG------CTGCAC---TGCTAAAAAATA-------------------------AT----ATACTA--CAAAACTT----------------------------------------------------------GCACAATA----C-----AA---------------ATAAAC------AAA--------------------------------------CA-------------------------------------------------------------CTT--AACACTGT---GT---------------------------------------------------------------------------------------------------------GAAAAACAGTCGCAGTTTCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droRho1 |
scf7180000779588:144466-144697 + |
|
CTGTACAA-------AA-ATAAC---G--TCATTTGTT------------------------------CA--------------------------------TA----------------------------TA-------T----------------------------GCCGTATCTCTT--TTC--TTTTT-----GCTCTTTTT------T----------------------------------------CTC----TCTCTTTCCGTGTTTTTAC---AAA--------------------ACTTCCTGTC----CG------CTGCAC---TGCTAAAAAAA--------------------------------ATACAA--CAAAACTT----------------------------------------------------------GCACAATA----C-----ACCG-------AAAAACCAAAAA------AAAAA------------------------------------C---------------------------A---------------------------------------AACACTGT---GT--------------------------------------------------------------------------------------------------------GAAAAAACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droBia1 |
scf7180000301345:418272-418503 - |
|
T-GTACAG-------A--ATAAT---G--TAATTCGTT----TAAACATATATAT------------ATATAT------------------------------------------------------------ATTCT----------------------------------------------TTT--TTTTT-----GTTCTCTTT------TTTCCTTC------TCT------------------------CTC----TCTCGTTCCGTGTTTTT------AA--------------------ACTTCCTGTC----CG------ATGCAC---CGCTAAAAAA----------------------------------------------ACT----------------------------------------------------------GCACCAAACACC---------A------------------------------------------------------------------------------------AAAAAAAATCCAACAAAAAAT------------------------------ACACTGT---CT-------------------------------------------------------------------------------------------------------GCAAAAAACAGTCGCAGTTCCGGTTCGACAACGAGTGCTATCTAAATCTGCCGGACAATCA |
| droTak1 |
scf7180000414531:38529-38766 - |
|
ATATATTA--------------C----------------------------ATATATA--TTTATATATATAT------------------------------------------------------------TTTCC----------------------------------------------TTT--TTTTTG----TCTC-TTTT------T----------------------------------------CTC----TCTCTTTCCGTGTTTTT--C--AAA--------------------ACTTCCTGTC----CG------ATGCAC---TGCTACAAAAA--------------------------AT--AAATACAAAT--------------------------------------------------------A--CATAAATGCACCACA----C-----ACCG-ACAACAAAA--------A--------AA---------------------------------------------------ACGAAAAATGTAT---------------------------------ACCCAC--AACACTGT---G---------------------------------------------------------------------------------------------------------AAAAAAACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTCAATCTGCCGGACAATCA |
| droEug1 |
scf7180000409182:27630-27902 + |
|
ATATATAC-------AA-AT-----------------------------ATATATATA--TTTATATTTATAT------------------------------------------------------------ATTCCTT--------------------------------------------TTCATTTTTT-----GCTCTTTTT------T----------------------------------------CTC----TCTCTTTCCGTGTTTTT--TA-CAA--------------------ACTTCCTGTC----CG------CTGCAC---TGCTATAAAA------------------------------------------------------------------TAA-AAACAAACAAACAAACAAAAAATCTATACAAAAAAATGCACAAA-----------------------AA--------AAA----A-A--------------------------------------CACAAAA--------------TTGTAC----------AACAAAAAAA---------AAAAAACGAAAAAACACTGT---GA--------------------------------------------------------------------------------------------------------AAAAAAACAGTCGCAGTTCCGTTTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| dm3 |
chrX:5119070-5119319 + |
|
ATATACTT---------------------------------TAAAACATATATATATA--TATATAAATATAT------------------------------------------------------------ATACATTA-------------------------------------------TTT--TTTTT-----GCTCTTTTT------T----------------------------------------CTCTC--TCTCTTTCCGTGTTTTT--T---AA--------------------ACTTCCTGTC----CG------CTGCAC---TGCTACAAAAA---------------------------------------------TTT----------------------------------------------------------GCACCACACACC---------C-ACA--------------C--------A--------------------------------------CACACACACACACAACGAACCATGTGCTGA----AA-AA-----------------------------CACACTGT---GA----A---------------AAT----------------------------------------------------------------------A--CA------AATAAAACAGTCCCAGTTCCGTTTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droSim2 |
x:4767467-4767701 + |
dsi_12387 |
ATATACTT---------------------------------TAAAACATATATATATA--CATATACATATAT------------------------------------------------------------ATACATT--------------------------------------------ATT--TTTTT-----GCTCTTTTC------T----------------------------------------CTCTC--TCTCTTTCCGTGTTTTT--T---GA--------------------ACTTCCTGTC----CG------CTGCAC---TGCTACAAAAA---------------------------------------------TTT----------------------------------------------------------GCACCACACACC---------ATGTGCTAAAA--------AAA----A-A--------------------------------------CACACACACAC---------------A-CAACAAAA-------------------------------------------------------------------------------AAT--------------------------------------------------------A--CA-----TAAAAAAACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droSec2 |
scaffold_0:3460922-3461078 + |
|
TCGCTCTC---------------------------------T---------CTCT---------------------------------------------------------------------------------------------------------------------------------CTC--TCTCT-----CTCTCTCTC------T------------------CTCTCTCTC--TTTCCCTCTTTCTC----TCTATCTCTGTCTGTTT--G--GGG--------------------GGCCTCTCTT----CG------CTCTGT---TGCAGCAGCGA-------------------------------------------------------------------AACAAAAAAT-------------------A--AATAAATAAATAACC----A-----GCTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAATAAAA--------------------------------------------------------GAAG-------AGAAAGAC---------------------------------------------------- |
| droYak3 |
X:3346217-3346440 - |
|
ACATATAT---------------------------------------------ATTTA--AATATATATATAT------------------------------------------------------------ATACA----------------------------------------------TTT--TTTTT-----GCTCTTTTT------T----------------------------------------CTC----TCTCTTTCCGTGTTCAT--T---GA--------------------ACTTCCTGTC----CGC-----CTGCAC---TGCTACAAAAA---------------------------------------------TTT----------------------------------------------------------GCACCACA----C-----AC---------------------------------------------------------------------------------AACAAACCATATGCCAA----AA-AA------------------------------ACACTGT---GA----A---------------AAT---GAAAAAAAAAA--------------------------------------------------------CTAC--------AAAATACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |
| droEre2 |
scaffold_4690:2474318-2474546 + |
|
ATATATAT-------A--------------------------------------TAGA--TATATATACATAT------------------------------------------------------------ATACATT---------------------------------------------AT--TTTTT-----GCTCTTTTT------T----------------------------------------CTC----TCTCTTTTCGTGTTTTT--T---AA--------------------ACTTCCTGTC----CG------CTGCAC---TGCAACAAAAA---------------------------------------------TTT----------------------------------------------------------GCACCACA----C-----AC---------------------------------------------------------------------------------AACAAACCATATGCCAA----AA---------------------------------ACATTGT---GA----A---------------AAT---AACAAAAAAAA--------------------------------------------------------ATACAAAAT-TCAAAAAACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTGAATCTGCCGGACAATCA |