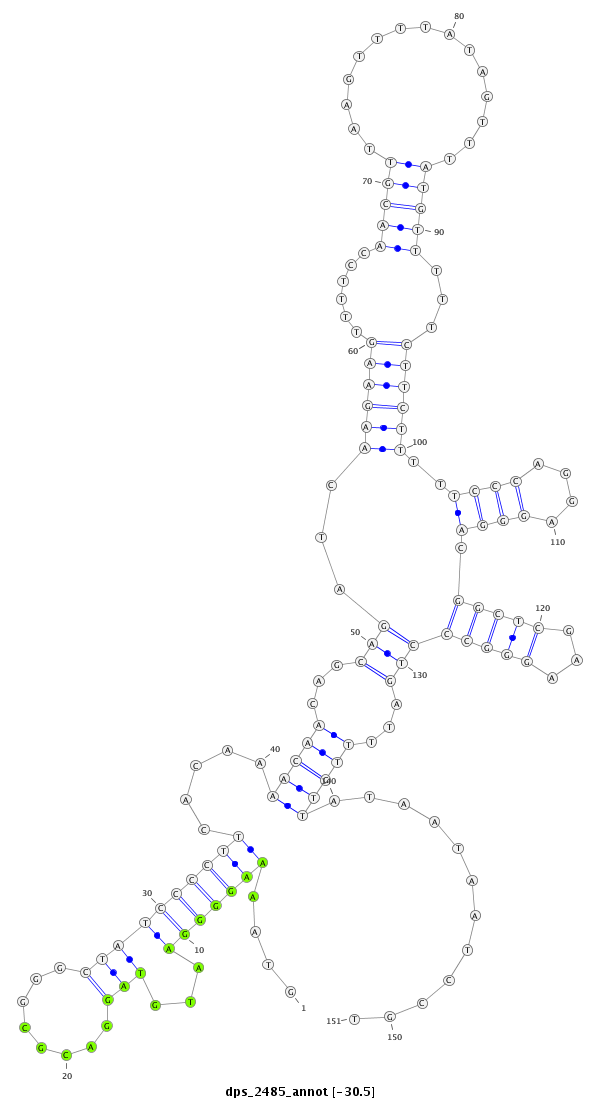
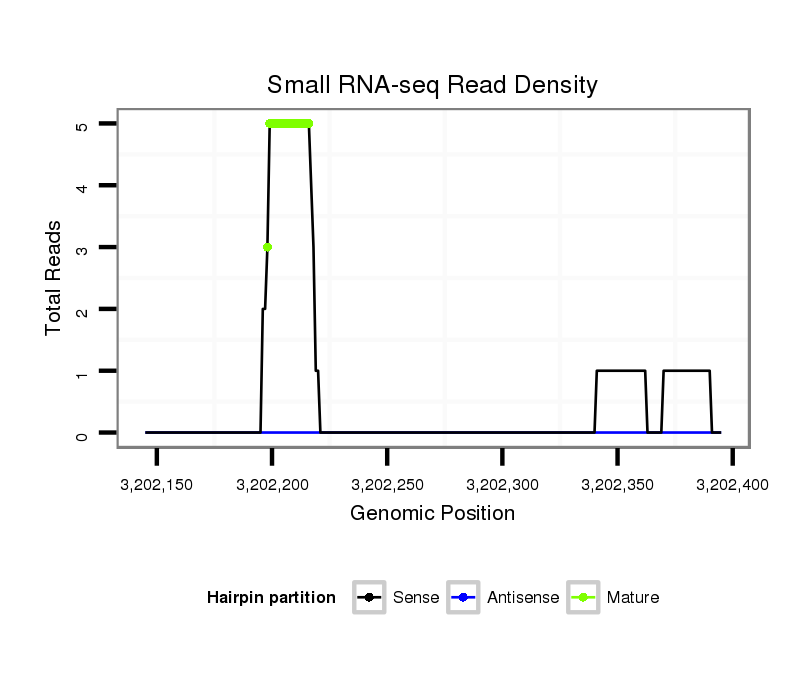
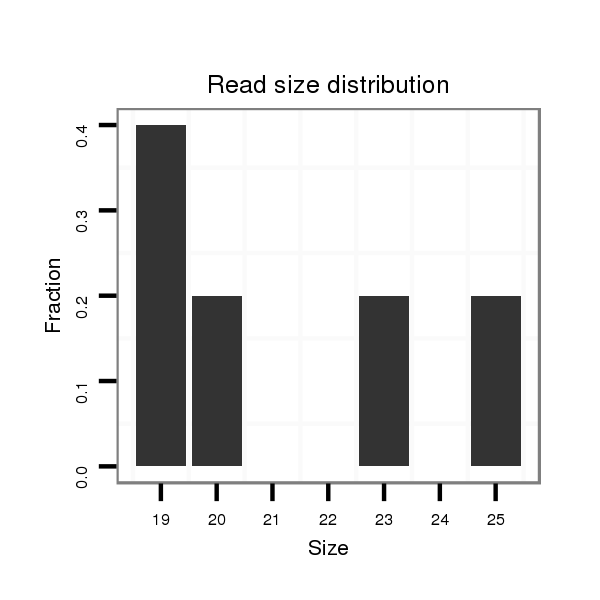
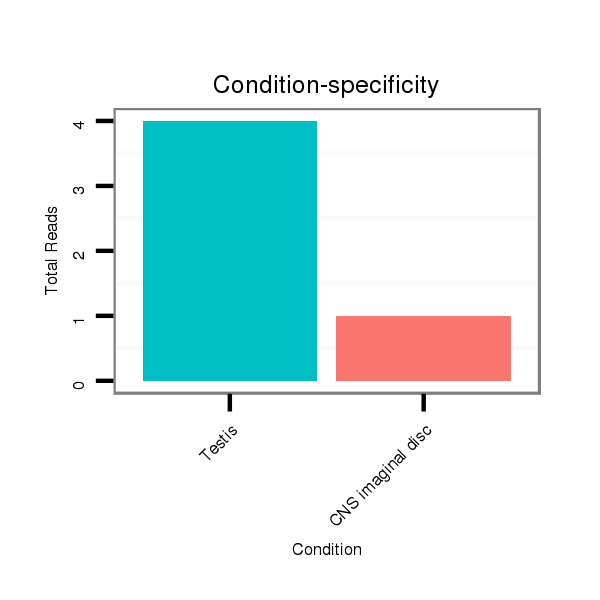
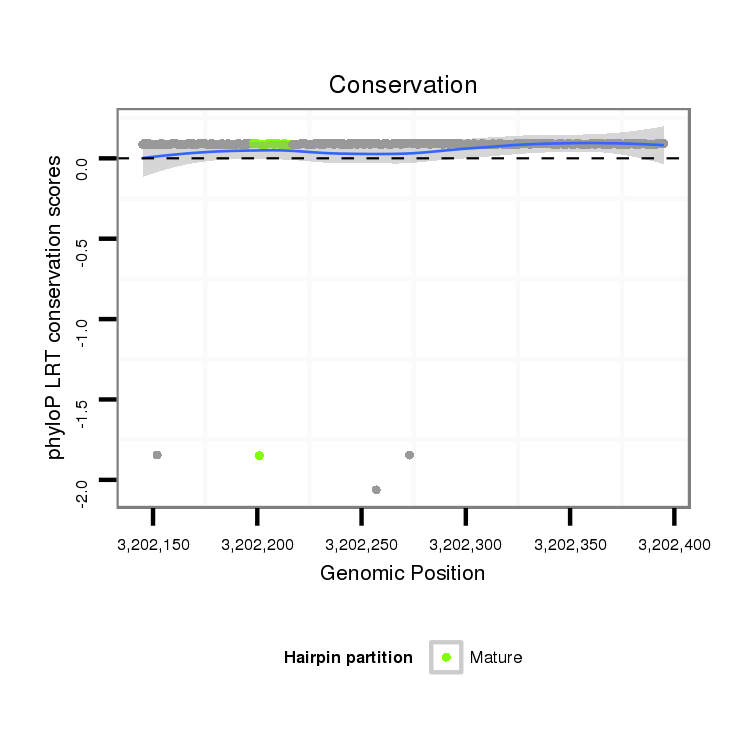
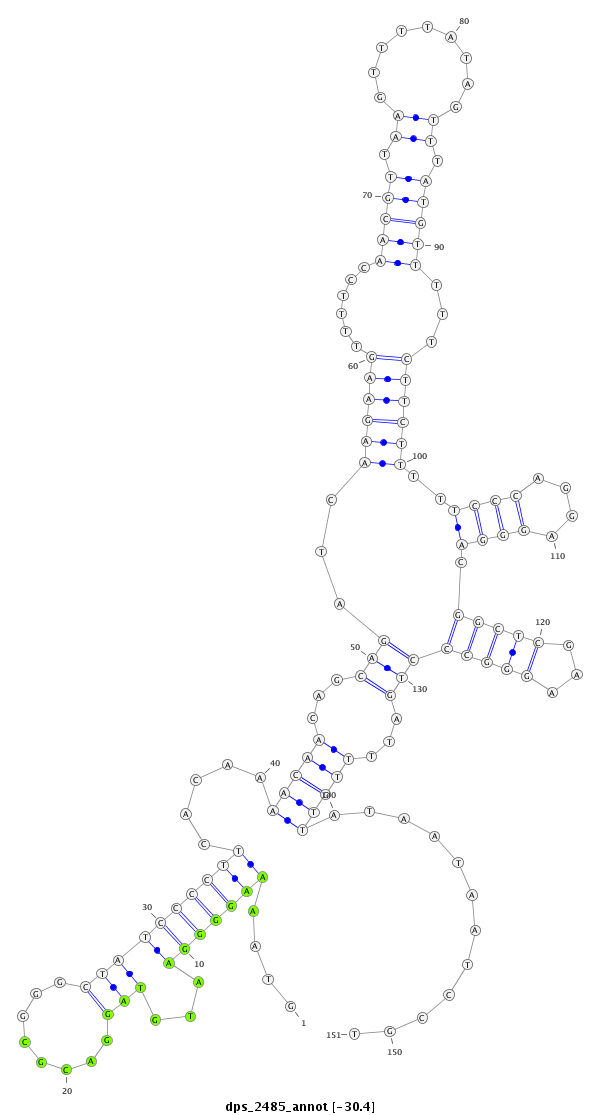
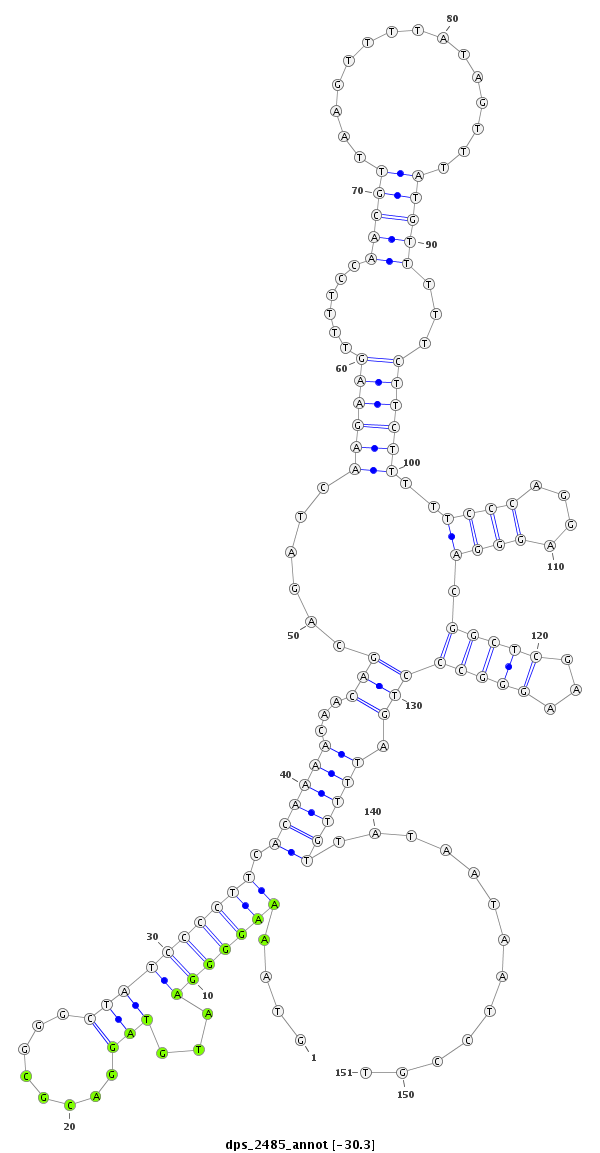
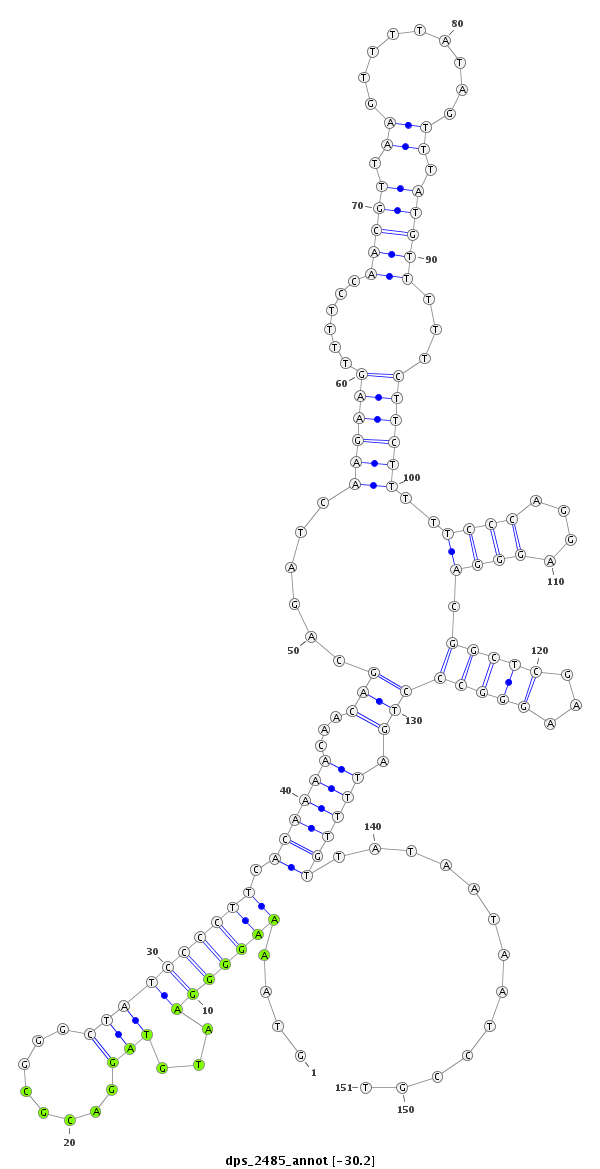
ID:dps_2485 |
Coordinate:XL_group1a:3202195-3202345 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -30.4 | -30.3 | -30.2 |
 |
 |
 |
CDS [Dpse\GA22958-cds]; exon [dpse_GLEANR_11338:1]; intron [Dpse\GA22958-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTCACCCTCAGCCGAAAGGGCGTCAGCATTTCGTCGCGTCCTCCCCAGCAGTAAAAGGGGAATGTAGGACGCGGGCTATCCCCTTCACAAAACAACAGCAGATCAAGAAGTTTTCCAACGTTAAGTTTTATAGTTTATGTTTTTCTTCTTTTTCCCAGGAGGGACGGCTCGAAGGGCCCTGATTTTGTTATAATAATCCGTAGGAAGCTTGGCCCTTGTTAGCTCTAAGTCCTGTACAGAAACCCCCATTA **************************************************....(((((((...(((........)))))))))).....(((((...(((...((((((......(((((...............)))))...))))))..((((....)))).(((((...))))))))...)))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
SRR902012 CNS imaginal disc |
GSM444067 head |
V112 male body |
GSM343916 embryo |
M022 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................AAAGGGGAATGTAGGACGC................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TAAAAGGGGAATGTAGGACGCGGGTTGG............................................................................................................................................................................ | 28 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TCCGTAGGAAGCTTGGCCCTTG................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TAAGTCCTGTACAGAAACCCC..... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAAAAGGGGAATGTAGGACGCGG................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................AAGGGGAATGTAGGACGCG.................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAAAAGGGGAATGTAGGACGCGGGC............................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................AAGGGGAATGTAGGACGCGG................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TGGAAGGGTCGTGATTTTGTT.............................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................ACAACAGGAGATCAAAGAGTT........................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................ACAACAGGAGATCAAGGAGCT........................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GTGTGGACGGCTCGAAGGGC.......................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................CAAGTGCAGAGCAAGAAGTT........................................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................GGACGAGTCTAAGGGCCC........................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GTTGGACGCGGGTTAGCC.......................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................GTTTGTGTTTATCTTCTTT.................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CAGTGGGAGTCGGCTTTCCCGCAGTCGTAAAGCAGCGCAGGAGGGGTCGTCATTTTCCCCTTACATCCTGCGCCCGATAGGGGAAGTGTTTTGTTGTCGTCTAGTTCTTCAAAAGGTTGCAATTCAAAATATCAAATACAAAAAGAAGAAAAAGGGTCCTCCCTGCCGAGCTTCCCGGGACTAAAACAATATTATTAGGCATCCTTCGAACCGGGAACAATCGAGATTCAGGACATGTCTTTGGGGGTAAT
**************************************************....(((((((...(((........)))))))))).....(((((...(((...((((((......(((((...............)))))...))))))..((((....)))).(((((...))))))))...)))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M062 head |
SRR902010 ovaries |
GSM343916 embryo |
GSM444067 head |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TCGAACCGGGACCATTCAAGA......................... | 21 | 3 | 1 | 19.00 | 19 | 18 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TCGAACCGGGACCATTCAAG.......................... | 20 | 3 | 5 | 8.40 | 42 | 42 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CGAACCGGGACCATTCAAGA......................... | 20 | 3 | 7 | 1.43 | 10 | 9 | 0 | 0 | 0 | 0 | 1 |
| ............................AAAGCAGTGCAGGAGGGGT............................................................................................................................................................................................................ | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TCGAACCGGGACCATTCAA........................... | 19 | 3 | 18 | 0.78 | 14 | 12 | 0 | 0 | 0 | 2 | 0 |
| .AGTGGGAGTCGGCGTACCC....................................................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TGTTGTCCTCTAGTTTCTCAA........................................................................................................................................... | 21 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CGGGAATAGTCGAGATTAAGGA.................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................AGGAGATGTGTTTGGGGGTGAT | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................GGAACAATCGAGAGGCAGGT.................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................GTCGAACCGGGACCATTCGA........................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TTGTTGTCCTCTAGTTTCTC............................................................................................................................................. | 20 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GTCGAACCGGGACCATTCG............................ | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................AAAAAGAAGGAAAGAGGTCCTC.......................................................................................... | 22 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................AAAAAATTATTAGTAGGCAT................................................. | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................GTTGTCCTCTAGTTTCTCAA........................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1a:3202145-3202395 + | dps_2485 | GTCACCCTCAGCCGAAAGGGCGTCAGCATTTCGTCGCGTCCTCCCCAGCAGTAAAAGGGGAATGTAGGACGCGGGCTATCCCCTTCACAAAACAACAGCAGATCAAGAAGTTTTCCAACGTTAAGTTTTATAGTTTATGTTTTTCTTCTTTTTCCCAGGAGGGACGGCTCGAAGGGCCCTGATTTTGTTATAATAATCCGTAGGAAGCTTGGCCCTTGTTAGCTCTAAGTCCTGTACAGAAACCCCCATTA |
| droPer2 | scaffold_12:13022-13272 - | GTCACCCACAGCCGAAAGGGCGTCAGCATTTCGTCGCGTCCTCCCCAGCAGTAAAAAGGGAATGTAGGACGCGGGCTATCCCCTTCACAAAACAACAGCAGATCAAGAAGTTGTCCAACGTTAAGTTTCATAGTTTATGTTTTTCTTCTTTTTCCCAGGAGGGACGGCTCGAAGGGCCCTGATTTTGTTATAATAATCCGTAGGAAGCTTGGCCCTTGTTAGCTCTAAGTCCTGTACAGAAACCCCCATTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 04:42 AM