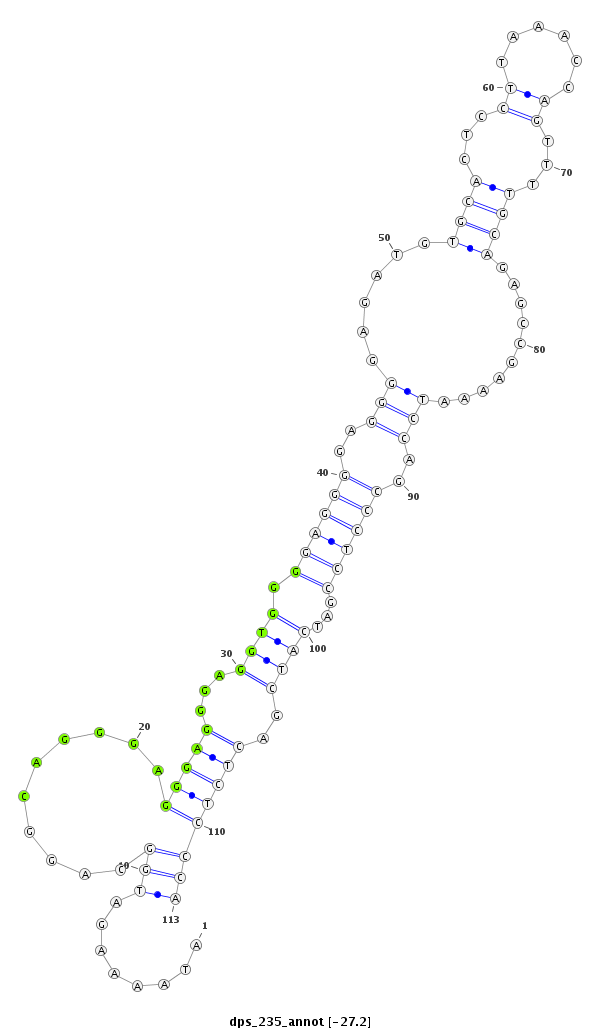
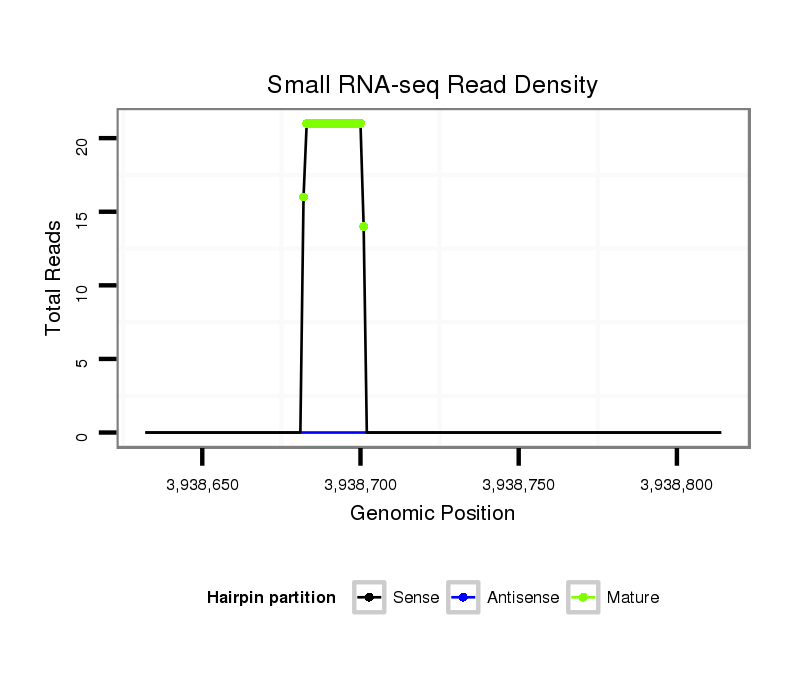

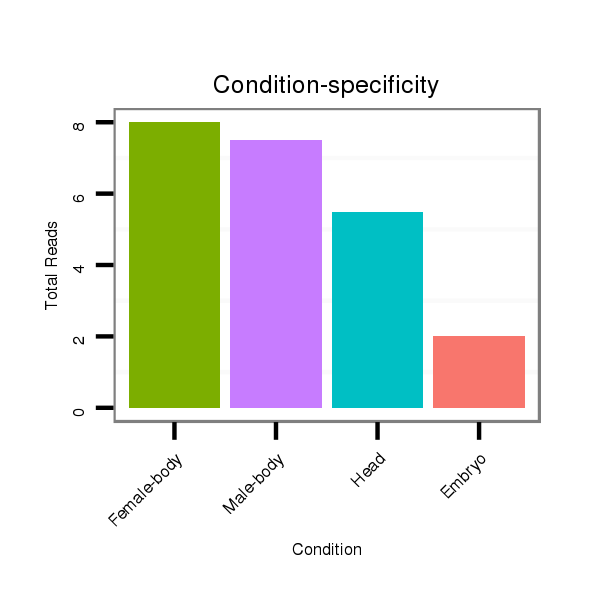
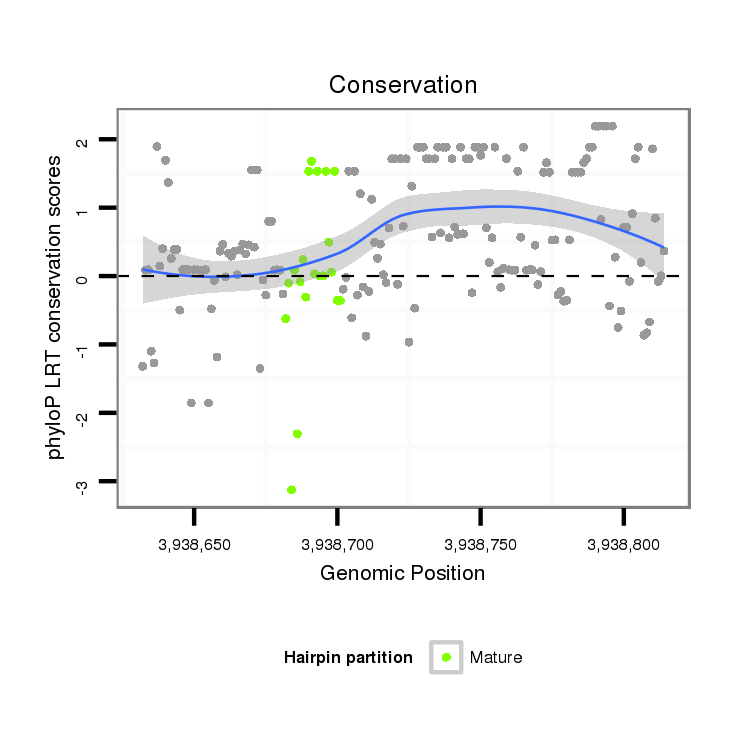
ID:dps_235 |
Coordinate:XR_group8:3938682-3938764 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -35.1 | -35.1 | -35.0 | -35.0 |
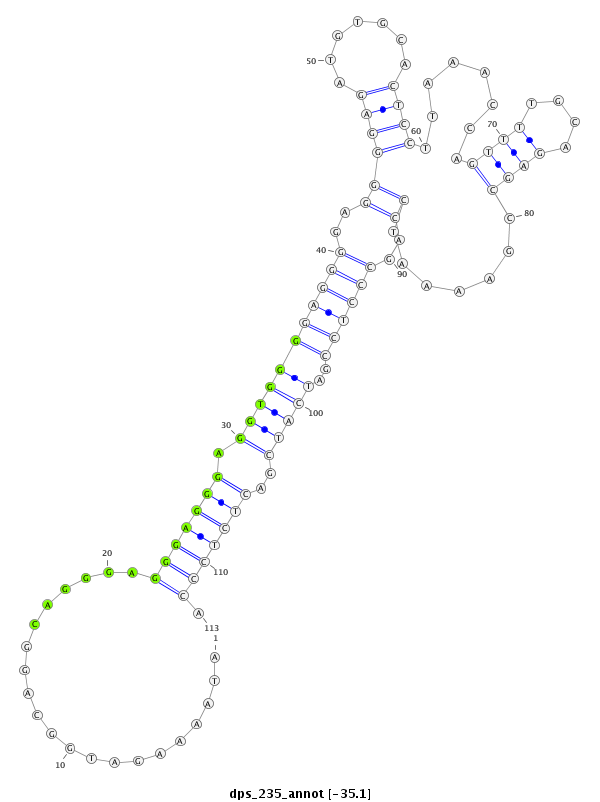 |
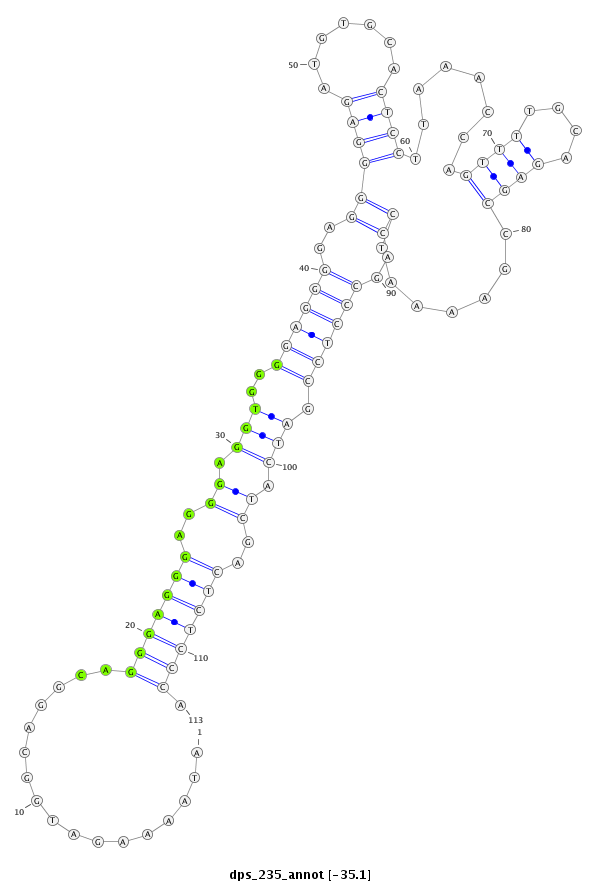 |
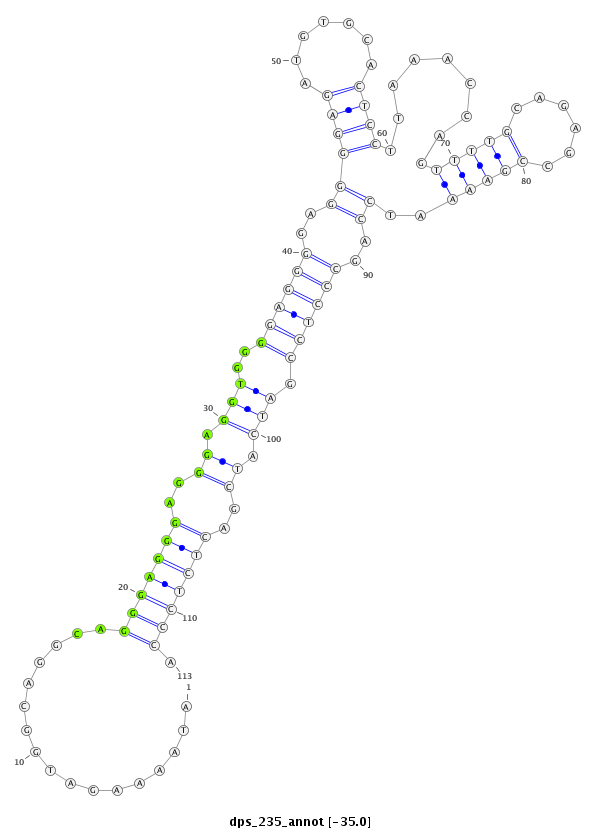 |
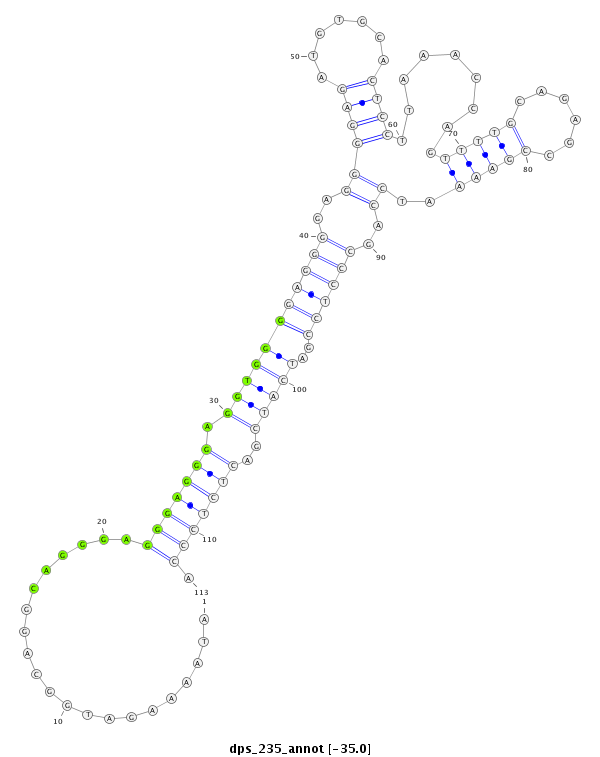 |
intron [Dpse\GA11027-in]
| Name | Class | Family | Strand |
| G-rich | Low_complexity | Low_complexity | + |
|
CCTATACCAAAAACAAAATACCAACAAAAATCCAGATAAAAGATGGCAGGCAGGGAGGGAGGGAGGTGGGGAGGGGAGGGGAGATGTGCACTCCTTAAACCAGTTTTGCAGAGCCGAAAATCCAGCCCTCCGATCATCGACTCTCCCATCGGCGTCAAAACAAGAAGAGGCACAACAACAATT
***********************************........(((..........(((((...((((.((((((..(((......((((...((......))...))))..........)))..))))))...))))..))))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
GSM444067 head |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................CAGGGAGGGAGGGAGGTGGG................................................................................................................. | 20 | 0 | 1 | 11.00 | 11 | 2 | 3 | 4 | 1 | 1 |
| ..................................................CAGGGAGGGAGGGAGGTGG.................................................................................................................. | 19 | 0 | 1 | 5.00 | 5 | 1 | 3 | 1 | 0 | 0 |
| ...................................................AGGGAGGGAGGGAGGTGGG................................................................................................................. | 19 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| ...................................................AGGGAGGGAGGGAGGTGG.................................................................................................................. | 18 | 0 | 2 | 2.00 | 4 | 3 | 1 | 0 | 0 | 0 |
| ........................................................GGGAGGAAGGTGGGGATG............................................................................................................. | 18 | 2 | 20 | 1.05 | 21 | 21 | 0 | 0 | 0 | 0 |
| ...................................ATAAAGGATGGCGGGCAGGG................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................GGAGGGAGGTGGGGATGA............................................................................................................ | 18 | 2 | 20 | 0.80 | 16 | 15 | 1 | 0 | 0 | 0 |
| ......................................................GAGGGAGGGAGGTGGGGA............................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................AGGGAGGTGGGGAGGGGA.......................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................GAGGGAGGTGGGGAGGGGAG......................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................GAGGTGGGGAGGGGAGGGG...................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................GAGGGAGTTGCGGAGGGGA.......................................................................................................... | 19 | 2 | 11 | 0.27 | 3 | 1 | 2 | 0 | 0 | 0 |
| ..................................................CAGGGAGGGGGGGAGGTGGG................................................................................................................. | 20 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CCGGGAGGGAGGGAGGTG................................................................................................................... | 18 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GCAGGGAGGTAGAGAGGGGAGGGG...................................................................................................... | 24 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................GCGAGGGAAGGAGGTGGGGA............................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TGGGAGGGAGGGAGGTGGG................................................................................................................. | 19 | 1 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................AAGATGGCAGGCACGGAAGT............................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GGCGGGAGGTGGGGAGGGG........................................................................................................... | 19 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................AGGGAGGGAGGGAGGTGGGA................................................................................................................ | 20 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................AGGGAGGAAGGTGGGGAT.............................................................................................................. | 18 | 2 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................AGGGAGGGAGGGAGGTTGGG................................................................................................................ | 20 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................CGGAGGGAGGTGGGGATG............................................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................GCCGGAGGGAGGTGGGGA............................................................................................................... | 18 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................AAGGGAGGGAGGTGGGGAGT............................................................................................................. | 20 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GGAGGGAGGTGGGGGGGGAAG......................................................................................................... | 21 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GGAGGTAGGGAGGGGAGGGGG..................................................................................................... | 21 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAGGGAGGTGGGAAGGGG........................................................................................................... | 19 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................GAGGGAGGTAGGAAGGGGA.......................................................................................................... | 19 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................GAGGGAGGGAGGTTGGGAG.............................................................................................................. | 19 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GGAGGAGGGGAGGGGAGGGGA..................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................AGGGAGGAGGGGAGGGGAGGGGCT.................................................................................................... | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................GAGGGAGGGAGGCGGGGA............................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................AAGGGAGGGAGGTGGGTA............................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GGATATGGTTTTTGTTTTATGGTTGTTTTTAGGTCTATTTTCTACCGTCCGTCCCTCCCTCCCTCCACCCCTCCCCTCCCCTCTACACGTGAGGAATTTGGTCAAAACGTCTCGGCTTTTAGGTCGGGAGGCTAGTAGCTGAGAGGGTAGCCGCAGTTTTGTTCTTCTCCGTGTTGTTGTTAA
***********************************........(((..........(((((...((((.((((((..(((......((((...((......))...))))..........)))..))))))...))))..))))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM444067 head |
V112 male body |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TCGGCTTATAGGTTGGCAGGC................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ...............................................................................................................TCGGCTTATAGGTGGGAAGGC................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................TAGTAGCTGATAGGGTAGTCGAAGTT......................... | 26 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................TGGCTGCAGTCTTGTTCTTC................ | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................................GCTGCAGTCTTGTTCTTCG............... | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 |
| ...........................................................................................AGGAATTGGGTAAAAAGGT......................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ............................................................................................GGAATTTGGTCGAGAGGTC........................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group8:3938632-3938814 + | dps_235 | CCTAT---------------ACCAAAAACAAAATACCAACAAAAATCCAGATAAAA----------------GATGGCAGGCAGGGAGG-GAG-GGAG-GTGGGGAG--GGGA-------------GG--GGAG---ATGTGCACTCCT-TAAACCAGTTTTGCAGAGCCGAAAATCCAGCCCTCCGATCATCGACTCTCC-CATCGGCGTCAAAAC--AAGAAGA-----------------------------------------------GG---------------------------C--ACAA--CAAC-------------------------AA-T--------T |
| droPer2 | scaffold_29:169779-169961 + | CCTTT---------------AGCAACAACAAAGTACCAGCAGAAATCG--------TA------GAT-AAAAGATGGCAGGCATGCAGG-GAGGGAGG-TAGGGAGGGGAGG------------------GGAG---ATGTGCACTCCT-TAAACCAGT-TTGCAGAGCCGAAAATCCAGCCCTCCGATCATCGACTCTCC-CATCGGCGTCAAAAC--AAGAAGA-----------------------------------------------GG---------------------------C--ACAA--CAAC-------------------------AA-T--------T | |
| droVir3 | scaffold_13049:120266-120275 + | A--AA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACC-------------------------AG-T--------T | |
| droMoj3 | scaffold_6680:1018457-1018478 - | A--AA---------------AAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAT--AAAATAA-----------------------------------------------AAC-------------------------------------------------------------------C--------A | |
| droBip1 | scf7180000395622:14737-14874 + | G--AG---------------AA---------------------------------------------------------------------------------------------------------------------AAGCCCTCTT-GAAACCAGT-TTTCAGAGCCAAAAAAAAAAAAC-----T---TGGCTCCT--CCATGGCGTCAAAAC--AACAGCT-------------------------------------------------C-CAGGAA---GACAACA--AAAACAG----CAGGAAGACAGGCAGTCCGGCAGTCAGGAACGGCAATA--------T | |
| droKik1 | scf7180000302577:1238433-1238664 + | A--TT---------------AACAACAAT----------CGAAATGGCTGAGAATA----------------A-------GCAT--TAA-GAAGGGGG-TTGAATGGGTTGA--------------------CT---GGATGCACTCTTTTAAACCAGTTTTGCTAAGCCCAAAATTCA-CCC---GAT---CAACTCTTTTTGGTGGCGTCAAAAC--AACAGACACAGAAATAATAACAACAAAAAGAACAGCAACAAAATGTGGAAGAACAAC-CAGAACAACAACAACA--ATATCAA----CAA--TAGC-------------------------AG-CAGCAGCTCT | |
| droFic1 | scf7180000454105:1385595-1385754 + | A------------------------------------------AAGCCTGGGAAAA----------------G-------ATAC--CGAAGAG-GTAG-TCGGCTGGGATGGAT-------------T--GGAT---GGCTGCTCTCCT-TAAACCCGT-TTGCAGAACCGAA-ATTCAGCCC---GAT---CAGCTCTT--TGGTGGCGTCAAAAC--AAGAGAG-----------------------------------------------AACCCAGAAC---TTGAACA--ATATCAA----CAT--CGTC-------------------------AG-C--------- | |
| droEle1 | scf7180000491249:2232961-2233132 - | A------------------------CAAA----------TGAAAAGCCTGAGAAAA----------------C-------GTAG--TGGAGAA-GGGG-TTGGATCGGCTGTAT----------------GGAT---GGATGCACTCCT-TAAACCCGT-TTGCAGAGCCGAAAATTCAGCCC---GAT---CAACTCTT--TGGTGGCGTCAAAAC--AACAGGG-----------------------------------------------AAC-CAGAAC---TACAACA--ATATCAAGCAACAA--AAGC-------------------------AA-C--------T | |
| droRho1 | scf7180000777063:191310-191486 - | A--T----------------AACAACAAT----------CGGAAAGCCTGAGAAAA----------------C-------ACTC--TGGAGAG-GGGG-TTGGCTGGGCTGG--------------------AT---GGATGCACTCCT-TAAACCCGT-TTGCAGAGCCGAAAATTCAGCCC---GGT---CAACTCTT--TGGTGGCGTCAAAAC--AACAGGG-----------------------------------------------AAC-CAGAACCACTACAACA--ATATCAACCAACAA--TAGC-------------------------TG-C--------T | |
| droBia1 | scf7180000302428:2413740-2413879 + | G---------------------------------------------------------------------------------------GAGAG-GGGG-TTGTCTGGGCTGGAT----------------GGA-TGTGGATGCGCACC--TAAACCCGT-TTGCAGAGCCGAAAATTCAGCCC---GAT---CAACTCTT--CGGTGGCGTCAAAAC--AACAGGC-----------------------------------------------AAC-CAGAAC---TGCGCCG--C-ATCAA----CAA--TGGC-------------------------AG-C--------T | |
| droTak1 | scf7180000415378:356256-356469 - | C--TAATACAAAGGTAATATAACAACGTT----------TGGAAAGCCTGAGAAAA----------------T-------ATTT--TGGAGAG-GGGGTTTGTCTGGGCAGGATGGATGGATGGCTGGATAGA-TATGGATGCACTCT--TAAACCCGT-TTGCAGAGCCGAAAATTCAGTCC---GAT---CAACTCTT--CGCTGGCGTCAAAACACAACAGGG-----------------------------------------------AAC-CAGAAC---TACAGCACAATATCAAC--ACAA--TAGC-------------------------AG-C--------T | |
| droEug1 | scf7180000409466:795007-795186 + | A--TA---------------AACAACAAT----------TGGAAAGCCTGAGAAAA----------------T-------ACAC--TGGAGAGGGGGG-TTGACTGGGAAGGATGGAT--------AT--AGA-TATGGATGCACTCT--AAAACCCGT-TTGCAGAGCCGAAAATTCAACTC---GAT---CCACTCTT--TAGTGGCGTCAAAAC--AACAGGG-----------------------------------------------AAC-CAGAAC---TACAACA--ATATCAA----CAA--TAG-----------------------------C--------C | |
| droSim2 | 3l:2932361-2932520 + | T--AT---------------AACAACAAC----------CAAGAGGCCTGAGTAAATT------GGT-AATAGGTGG-----------CAGAG-GGGG-TTGGCTGGGCTGA--------------------AT---GAGTGCACTCG--CAAACCCGT-TTGCAGAGCCCAAAATTCAGCCC---GAT----------------------CAAAAC--AACAGCG-----------------------------------------------AAC-CAGAAC---TGCAGCG--CTATCAA----CAA--TGGC-------------------------AG-C--------T | |
| droSec2 | scaffold_2:3068033-3068199 + | T--AT---------------AACAACAAC----------CAAGAGGCCTGAGTAAATTGATATAGGTAAATAGGTGG-----------CAGAG-GGGG-TTGGCTGGGCTGA--------------------AT---GAATGCACTCG--CAAACCCGT-TTGCAGAGCCCAAAATTCAGCCC---GAT----------------------CAAAAC--AACAGCG-----------------------------------------------AAC-CAGAAC---TGCAGCG--CTATCAA----CAA--TGGC-------------------------AG-C--------T | |
| droYak3 | 3L:8872767-8872938 - | T--ACCTG-AAAGGG--TATAACAACAAC----------CAAGCAGCCCAAGTAAATT------GGC-AATAGGTGG-----------CAGAG-GGGG-TTGGCTGGGCTGA--------------------AT---GAATGCACTCG--CAAACCCGT-TTGCAGAGCCGAAAATTCAGACC---GAT----------------------CAAAAC--AACAGCG-----------------------------------------------AAC-CAGAAC---TTCAACG--CTATCAA----CAA--TAGC-------------------------AG-C--------T | |
| droEre2 | scaffold_4784:3082251-3082379 + | G--G--------------------------------------------------------------T-AAAAGGCGG-----------CAGAG-GGGG-TTGGCCGGGTTGA--------------------AT---GAATGCACTCG--TAAACCCGT-TTGCAGAGCCGAAAATTCAGCCC---GAT----------------------CAAAAC--AAAAGCG-----------------------------------------------AAC-CAGAAC---TACAGCG--CTATCAA----CAA--AGCC-------------------------AG-C--------T |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/15/2015 at 03:11 PM