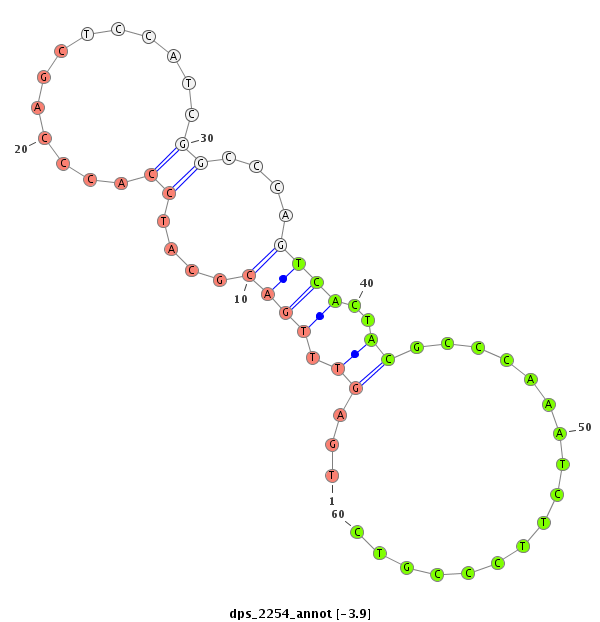
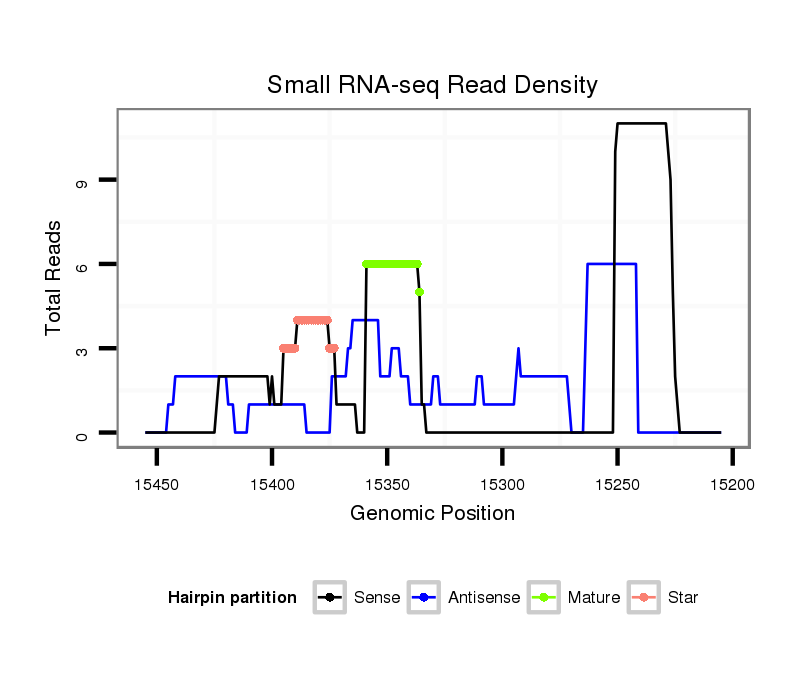
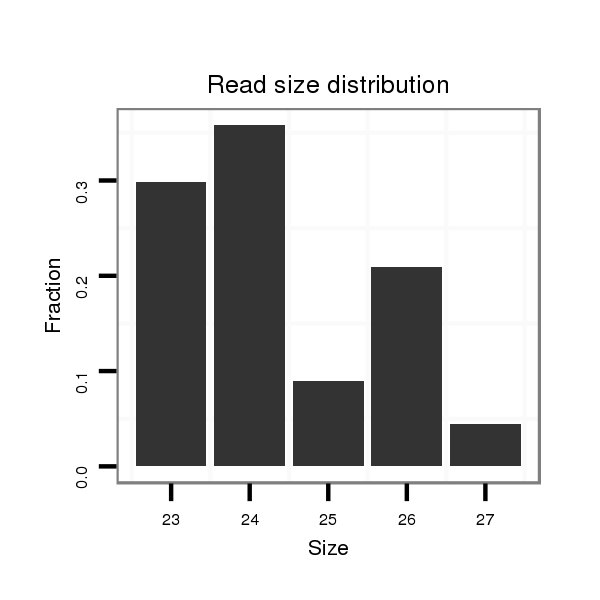
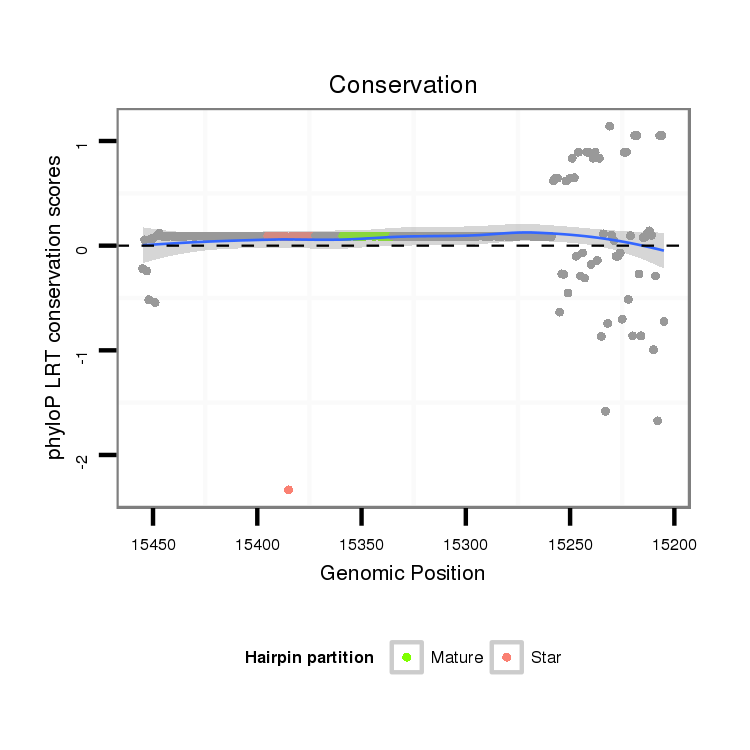
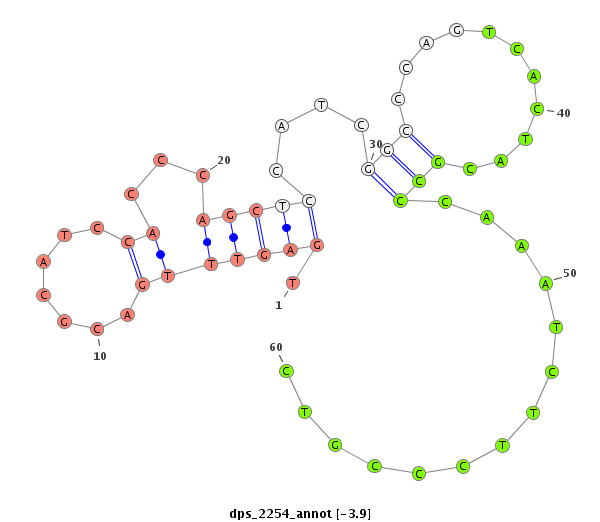
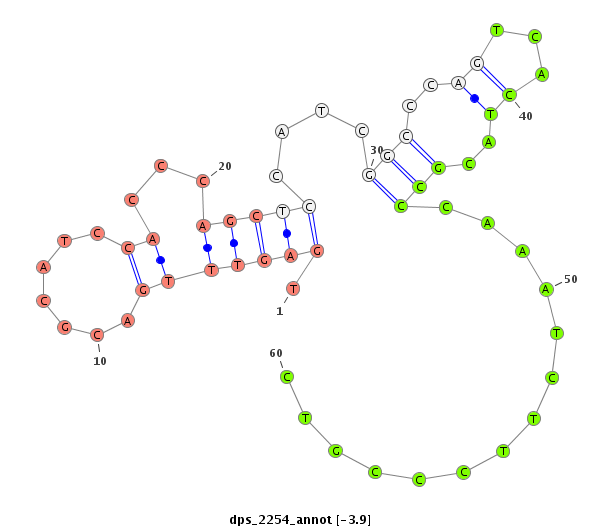
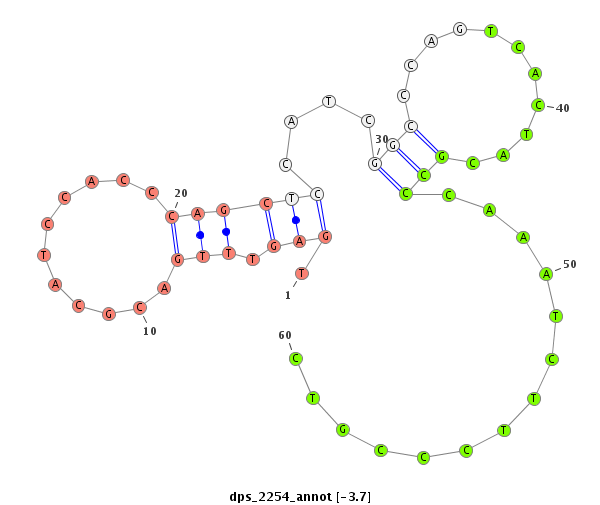
| ................................................................................................................................................................................................GGAGCCATCAGGGAGCGTGCCA..................................... |
22 |
0 |
4 |
3.00 |
12 |
12 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................GGGAGCCATCAGGGAGCGTGCCA..................................... |
23 |
0 |
4 |
3.00 |
12 |
11 |
1 |
0 |
0 |
0 |
| .................................................................................CGAGGTAGCCGGGTCAGTGAT..................................................................................................................................................... |
21 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..........................................................................................CGGGTCAGTGATGCGGGTTTAGAAG........................................................................................................................................ |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| .............................................TCGCCCACTCAAGCGACTCAAACTG..................................................................................................................................................................................... |
25 |
0 |
4 |
1.00 |
4 |
1 |
0 |
0 |
3 |
0 |
| .................................................................................................................................................................TTTGGTGTACGGAAGCTCCGAAAT.................................................................. |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................................................TTTAGAAGGGCAGACCCGGGT........................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........CGGTTGACCACTTTTCTTTAGAGCGT....................................................................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................TTGGTGTACGGAAGCTCCGAAA................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................CGTGCAGGGCCCGAGTGTT........................................................................................ |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............TTGACCACTTTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................................................................................................................GGTCGACTCGCTCCAGGCTCGT........................................................................................................ |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................GCCGGGTCAGTGATGCGGGTTTA............................................................................................................................................ |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................GGGAGCCATCAGGGAGCGTGCCAA.................................... |
24 |
0 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..............................................CGCCCACTCAAGCGACTCAA......................................................................................................................................................................................... |
20 |
0 |
8 |
0.63 |
5 |
5 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................GAGCCATCAGGGAGCGTGCCA..................................... |
21 |
0 |
5 |
0.60 |
3 |
2 |
1 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................AAATTAGCCGGCGGTCAAGGCGT |
23 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................AAGGGCAGACCCGGGTCGACTCGT................................................................................................................... |
24 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............................................CGCCCACTCAAGCGACTCAAACTG..................................................................................................................................................................................... |
24 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...............GACCACTGTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
24 |
1 |
12 |
0.50 |
6 |
6 |
0 |
0 |
0 |
0 |
| ...............................................GCCCACTCAAGCGACTCAA......................................................................................................................................................................................... |
19 |
0 |
8 |
0.50 |
4 |
4 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................GGGAGCCCTCAGGGAGCGTGCCA..................................... |
23 |
1 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CCCGGGTCGACTCGCTCCAG.............................................................................................................. |
20 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AGCCATCAGGGAGCGTGCCAA.................................... |
21 |
0 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| .................................................................................................................AGGGCAGACCCGGGTCGACTCG.................................................................................................................... |
22 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AGCCCTCAGGGAGCGTGCCA..................................... |
20 |
1 |
10 |
0.30 |
3 |
3 |
0 |
0 |
0 |
0 |
| ................ACCACTTTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
23 |
0 |
7 |
0.29 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...............GACCACTTTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
24 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................AGGGGAGCCATCAGGGAGCGTGCCA..................................... |
25 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GGAGCCATCAGGGAGCGTGCCAA.................................... |
23 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGGGAGCCATCAGGGAGCGTGCCA..................................... |
24 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................AGGGGAGCCATCAGGGAGCG.......................................... |
20 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GGAGCCATCAGGGAGCGTGCC...................................... |
21 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................AGAGCCATCAGGGAGCGTGCCAA.................................... |
23 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................GAGCCATCAGGGAGCGTGC....................................... |
19 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................GAGCCATCAGGGAGCGTGCCAAG................................... |
23 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................TCAGGGAGCGTGCCAAGGATCGT............................. |
23 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AGCCATCAGGGAGCGTGCCAAGG.................................. |
23 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................TCGTGCACGGCTCGTGTGTT........................................................................................ |
20 |
3 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
1 |
| ..................................................................................................................................................................................................AGCCATCAGGGAGCGTGCCA..................................... |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................GAGCCATCGGGGAGCGTGCCA..................................... |
21 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................GCCATCAGGGAGCGTGCCAA.................................... |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................................TCGCCCACTCAAGCGACTCAA......................................................................................................................................................................................... |
21 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................AAGGATCGTGGTATCGTTTT.................. |
20 |
3 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GGAGCCCTCAGGGAGCGTGCCAA.................................... |
23 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GGAGCCCTCAGGGAGCGTGCCA..................................... |
22 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................CCACTGTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
22 |
1 |
15 |
0.13 |
2 |
2 |
0 |
0 |
0 |
0 |
| ................ACCACTGTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
23 |
1 |
15 |
0.13 |
2 |
2 |
0 |
0 |
0 |
0 |
| ................ACCACTTTTCTTTAGAGCGTTC..................................................................................................................................................................................................................... |
22 |
0 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................CGACTTTTATTTAGAGCATTCTTC.................................................................................................................................................................................................................. |
24 |
3 |
9 |
0.11 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................TCAGGGAGCGTGCCAAGACTC............................... |
21 |
2 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GGAGCCCTCAGGGAGCGTGCCAAGACT................................ |
27 |
3 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............TGACCACTGTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
25 |
1 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............AGACCACTGTTCTTTAGAGCGTTCT.................................................................................................................................................................................................................... |
25 |
2 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............GACCACTGTTCTTTAGAGCGTTCTT................................................................................................................................................................................................................... |
25 |
1 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............GACCACTGTTCTTTAGAGCGTTC..................................................................................................................................................................................................................... |
23 |
1 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............TGACCACTGTTCTTTAGAGCGT....................................................................................................................................................................................................................... |
22 |
1 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................GAGCCCTCAGGGAGCGTGCCAAGACT................................ |
26 |
3 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................GTCGACTCGCTCCAGGCT........................................................................................................... |
18 |
0 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AGCCCTCAGGGAGCGTGCCAAGACTCGT............................. |
28 |
3 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................CCACTGTTCTTTAGAGCGTTCTTC.................................................................................................................................................................................................................. |
24 |
1 |
15 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AGCCCTCAGGGAGCGTGCCAAGACT................................ |
25 |
3 |
15 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................................................ATAGCCGGCGGTCAAGGCGT |
20 |
1 |
16 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................TGGTGTACGGAAGGTGTGA..................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
| ..................AACTGTTCTTTAGAGCGTTCTTC.................................................................................................................................................................................................................. |
23 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................GCCAACTCAACCGACTCAG......................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................AAGTCAAGCGATTCAAACT...................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |