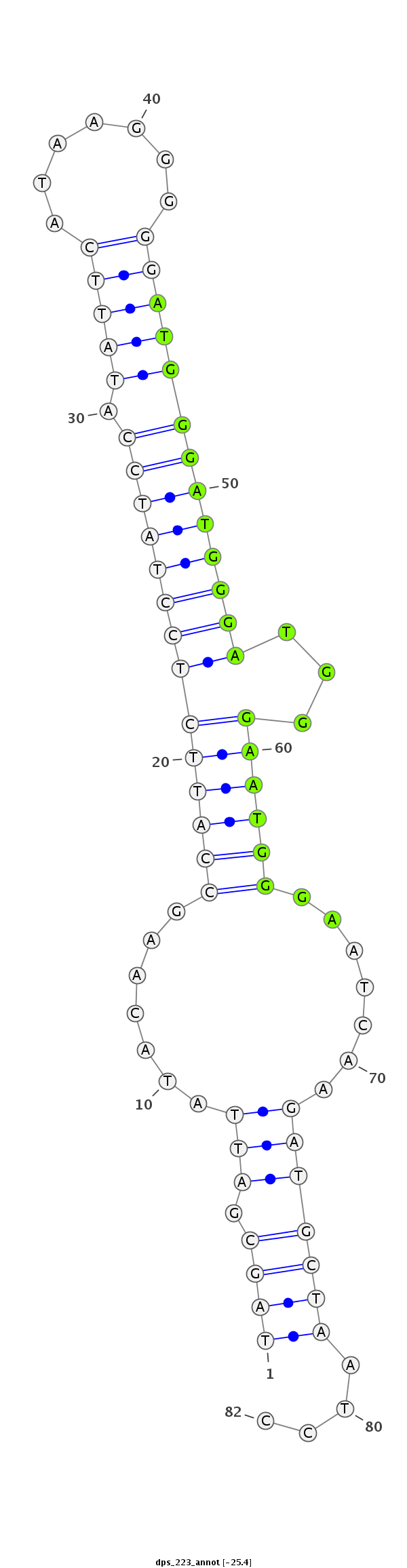


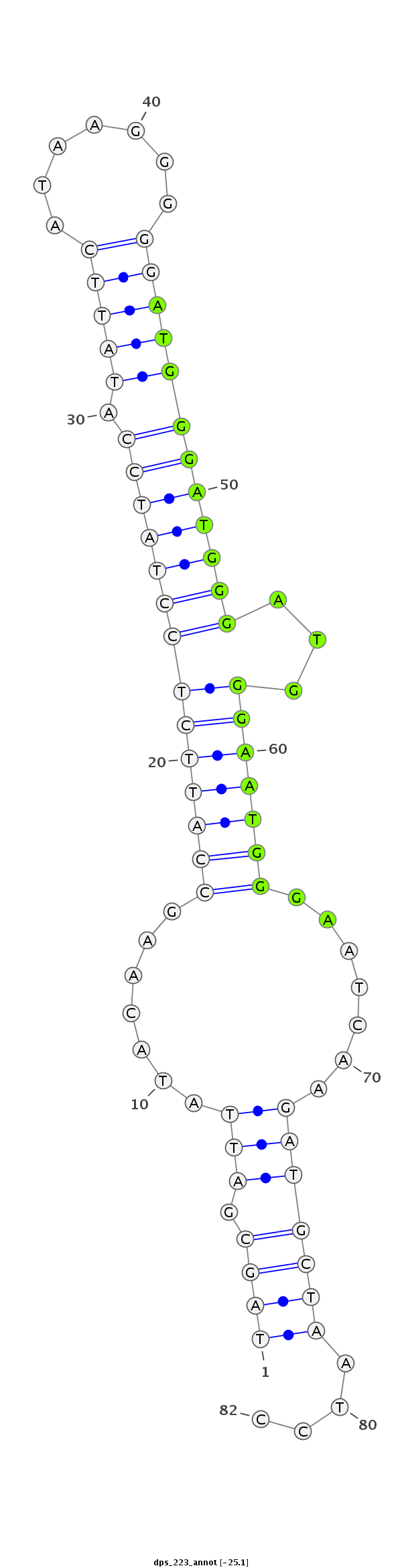
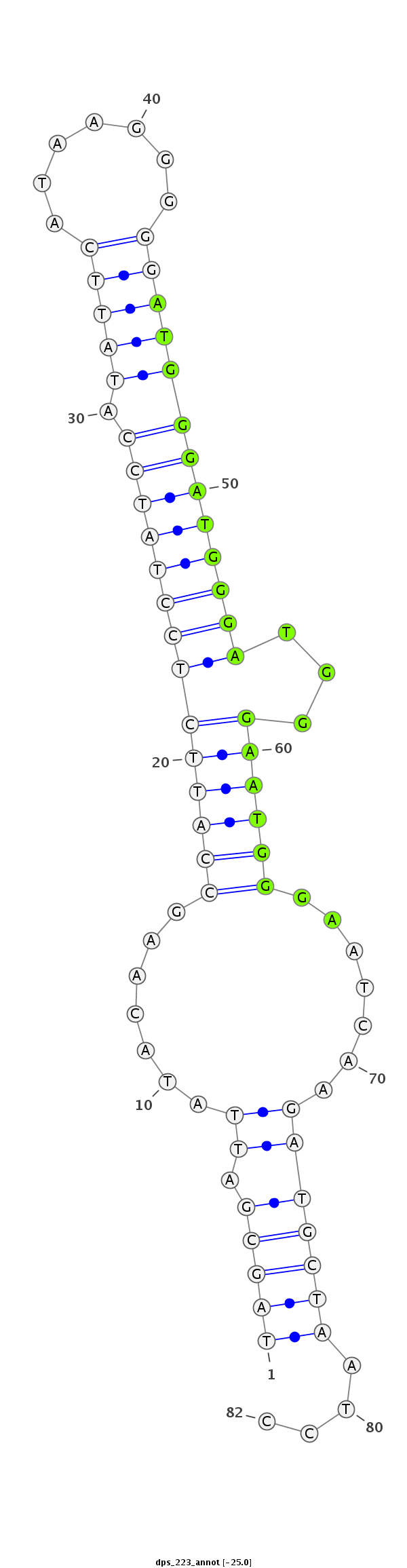
ID:dps_223 |
Coordinate:XL_group1a:3362787-3362838 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -25.2 | -25.1 | -25.0 |
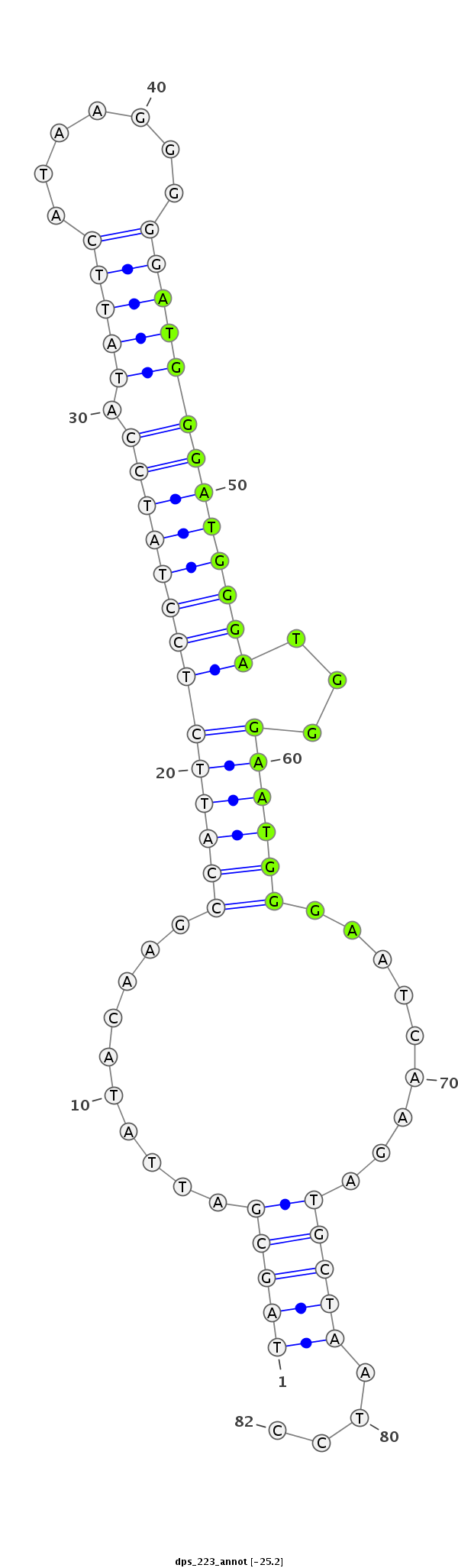 |
 |
 |
Antisense to intron [Dpse\GA17935-in]
No Repeatable elements found
| mature | star |
|
AAATATTAGCAGGTGGAAGGTTGGGGGAATAACATTAGCGATTATACAAGCCATTCTCCTATCCATATTCATAAGGGGGATGGGATGGGATGGGAATGGGAATCAAGATGCTAATCCCCAGGTTCCAGGCTGCCAGGCGTCCAGGTCTCGAG
***********************************((((.(((.......((((((((((((((.(((((.......)))))))))))))...)))))).......)))))))....*********************************** |
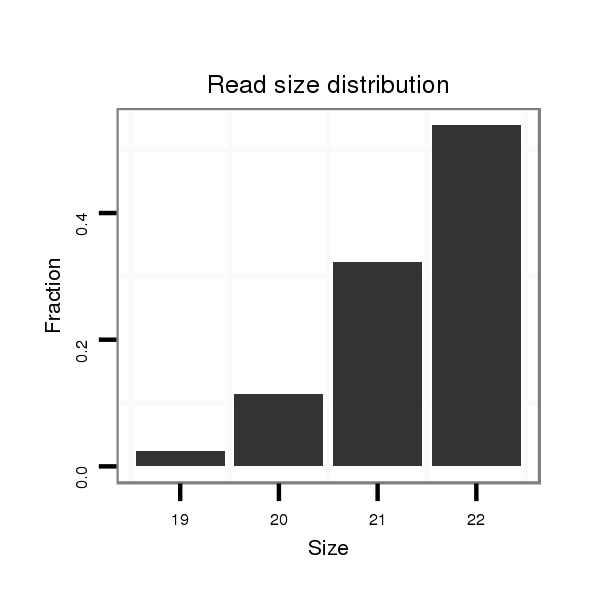
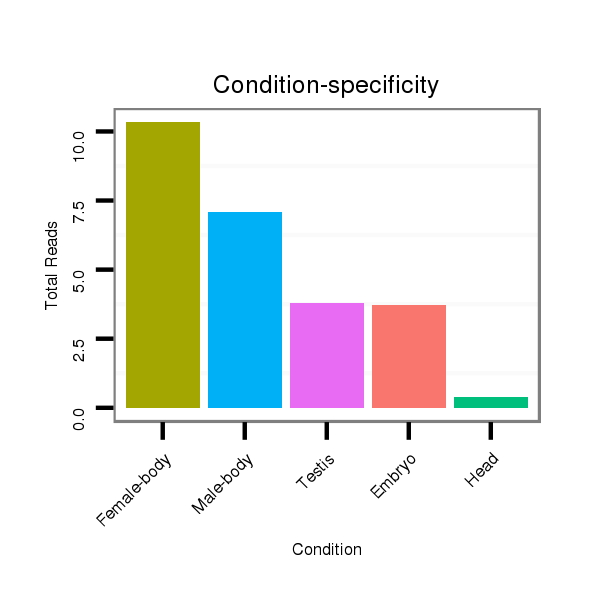
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
SRR902011 testis |
GSM343916 embryo |
M059 embryo |
GSM444067 head |
SRR902012 CNS imaginal disc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................ATGGGATGGGATGGGAATGGGA................................................... | 22 | 0 | 3 | 13.67 | 41 | 17 | 12 | 5 | 6 | 1 | 0 | 0 |
| ...............................................................................ATGGGATGGGATGGGAATGGG.................................................... | 21 | 0 | 5 | 5.60 | 28 | 7 | 11 | 3 | 4 | 1 | 2 | 0 |
| ................................................................................TGGGATGGGATGGGAATGGG.................................................... | 20 | 0 | 5 | 2.40 | 12 | 3 | 2 | 5 | 1 | 1 | 0 | 0 |
| ................................................................................TGGGATGGGATGGGAATGGGA................................................... | 21 | 0 | 3 | 2.33 | 7 | 5 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........GGTGGAAGTTTGAGGCAATAACA...................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TAAGGGTGAGGGGATGGGATG............................................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GATGGAAGTTTGGGGCAATAACA...................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......AGCCGGAGGAAGGTTGGG............................................................................................................................... | 18 | 2 | 8 | 0.63 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....ATGAGCAGGTTGAAGGTTGG................................................................................................................................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................GGGTGTGAAGGGATGGGAATGGGAA.................................................. | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GATGGAAGTTTGGGGCAATAAC....................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAGGGTGCCAGGCGTCGATGT...... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................TGGGATGGGATGGGAATGG..................................................... | 19 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GATGGAAGGGGATTGGGAATCA............................................... | 22 | 3 | 7 | 0.29 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................GGGATGGGATGGGAATGGGA................................................... | 20 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TGGGATGGGATGGGAATGGGAT.................................................. | 22 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................GGGGGATGGGATGGGATGGGA......................................................... | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GGTGGAAGTTTGAGGCAATAA........................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGGGATGGGATGGGATGGG.......................................................... | 19 | 0 | 20 | 0.20 | 4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................ATGGGATGGGATGGGAATGG..................................................... | 20 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............GTGGAAGGTCGTGGGAAT.......................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................ATGGGATGGGATGGGAATGGGC................................................... | 22 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................AGGGGTGTGGGATGGGATGGGA......................................................... | 22 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................TGGGATGGGATGGGAGTGGG.................................................... | 20 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GGAAGTTTGAGGTAATAACA...................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......AAGCCGGAGGAAGGTTGGG............................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................AAGGGGGATAGTGTGGGATG............................................................ | 20 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGGGATGGGATGGGATGGGA......................................................... | 20 | 0 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................GGATGTGGTGGGATGGGAAAGG..................................................... | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......AGCCGGTGGAAGGTGGGGA.............................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CAGGGGATGGGATGGGAAT....................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
TTTATAATCGTCCACCTTCCAACCCCCTTATTGTAATCGCTAATATGTTCGGTAAGAGGATAGGTATAAGTATTCCCCCTACCCTACCCTACCCTTACCCTTAGTTCTACGATTAGGGGTCCAAGGTCCGACGGTCCGCAGGTCCAGAGCTC
***********************************((((.(((.......((((((((((((((.(((((.......)))))))))))))...)))))).......)))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM444067 head |
M040 female body |
|---|---|---|---|---|---|---|---|
| ................................................TCGGTAAGAGGACAGGCATAAT.................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 |
| .......................CCCCTTATTGTAATCGCTAATA........................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| .................................................AGGTAAGAGGACAGGTATAAT.................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| .................................................AGGTAAGAGGAAAGGTATAAT.................................................................................. | 21 | 3 | 18 | 0.06 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1a:3362737-3362888 - | dps_223 | AAATATTAGCAGGTGGAAGGTTGGGGGAATAACATTAGCGATTATA-CAAGCCATTCTCCTATCCATATTCA--TAAGGGGGATGGGATGGGA----TGGGAATGGGAATCAAGATGCTAATCCCCAGGTTCCAGGCTGCCAGGCGTCCAGGTCTCGAG |
| droPer2 | scaffold_9:1378075-1378108 + | GGA-------------------------------------------------------------------------------ATGGGATGGGA----TTGGGATGGGGATGGGGATG------------------------------------------ | |
| droMoj3 | scaffold_6359:1625998-1626035 - | GACTACTGGAAGGGGG------GGGGGAATAACATTAGCAATTA------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000414425:241891-241975 - | GAC------------------TGGGGGAATAACATTAGCAATTAAAGCGGGCTATTTT-------ACATTTAGCTA---GGGATGGCAAGGGAATACTTGGAATGATGATCGA---------------------------------------------- | |
| droEug1 | scf7180000409110:954609-954640 - | GAC------------------TGGGGGAATAACATTAGCAATTAAA----GCCA--------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/15/2015 at 03:10 PM