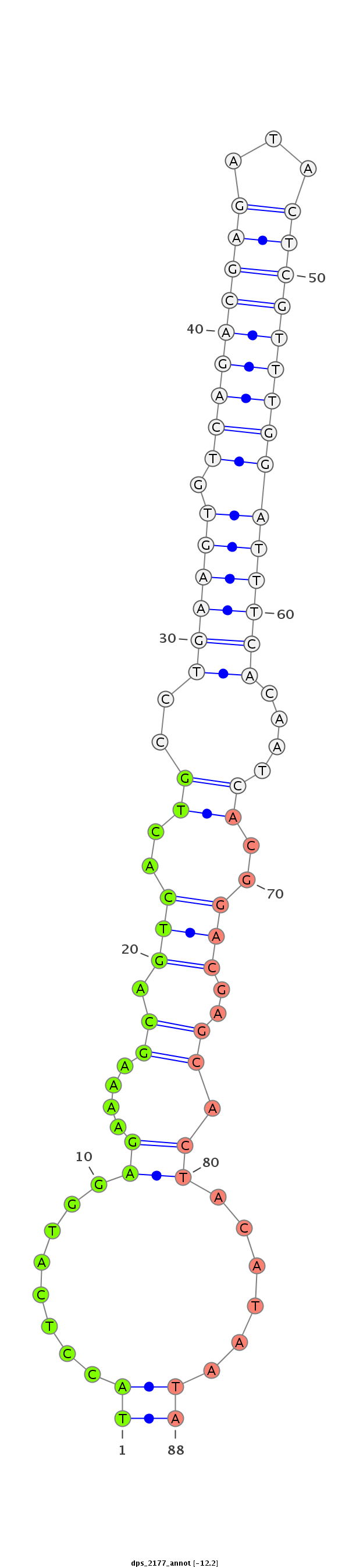
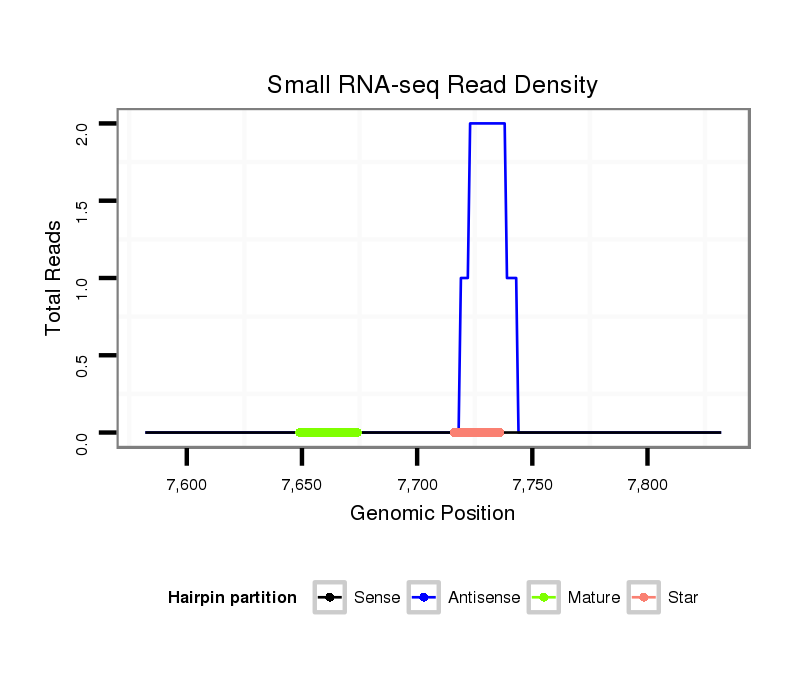
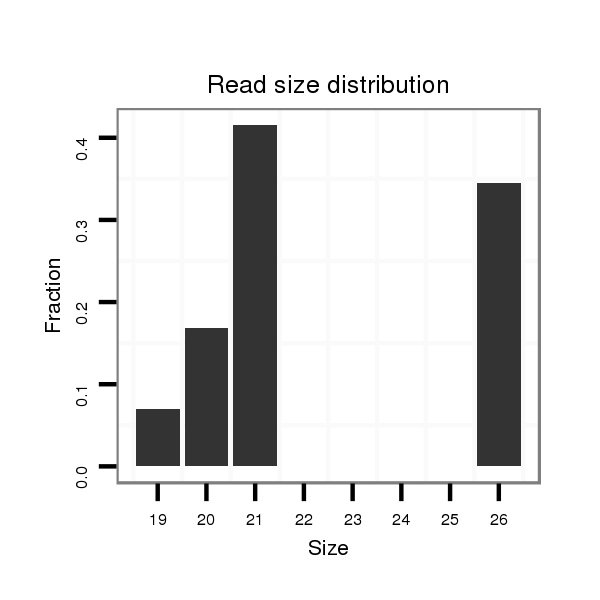
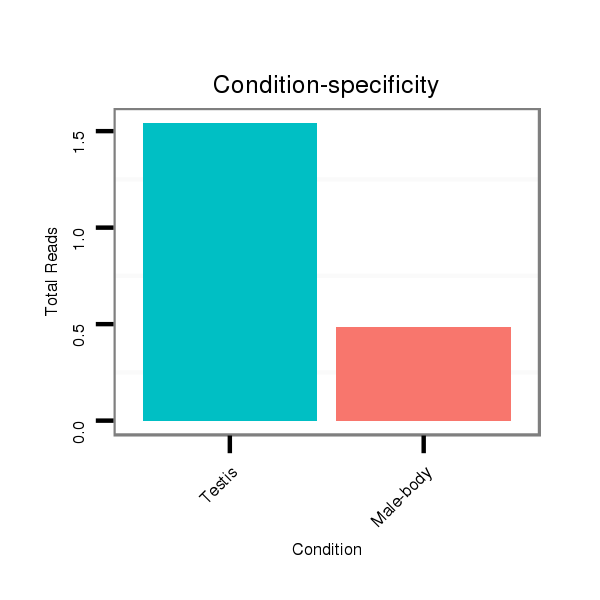
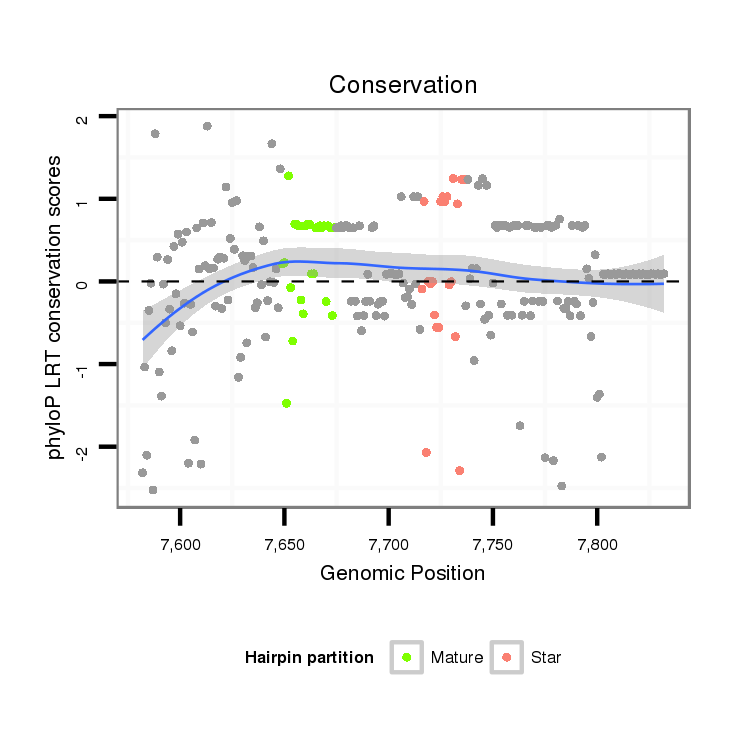
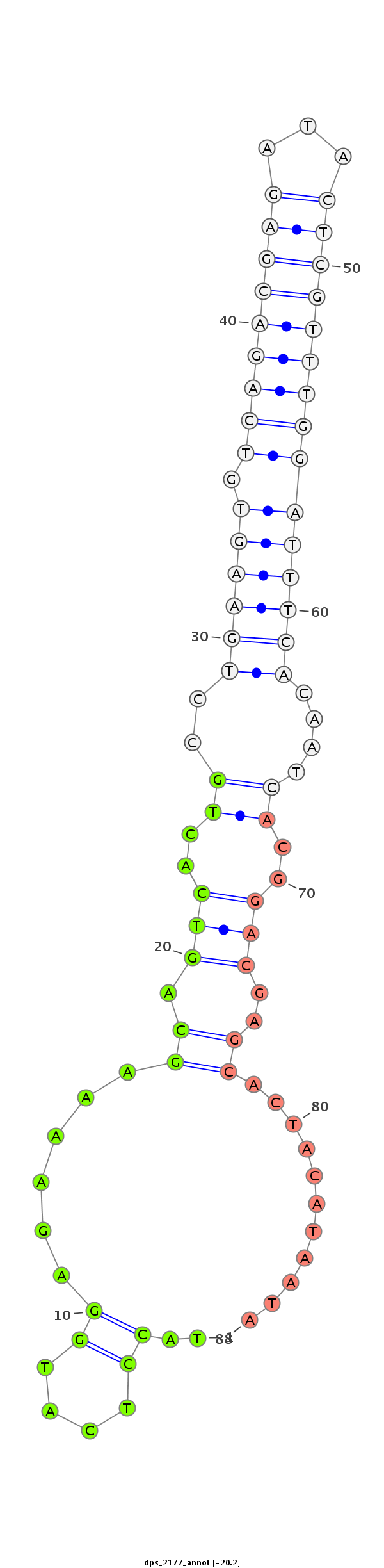
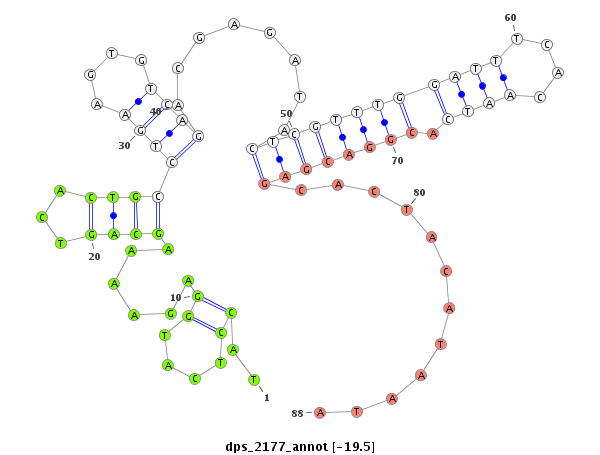
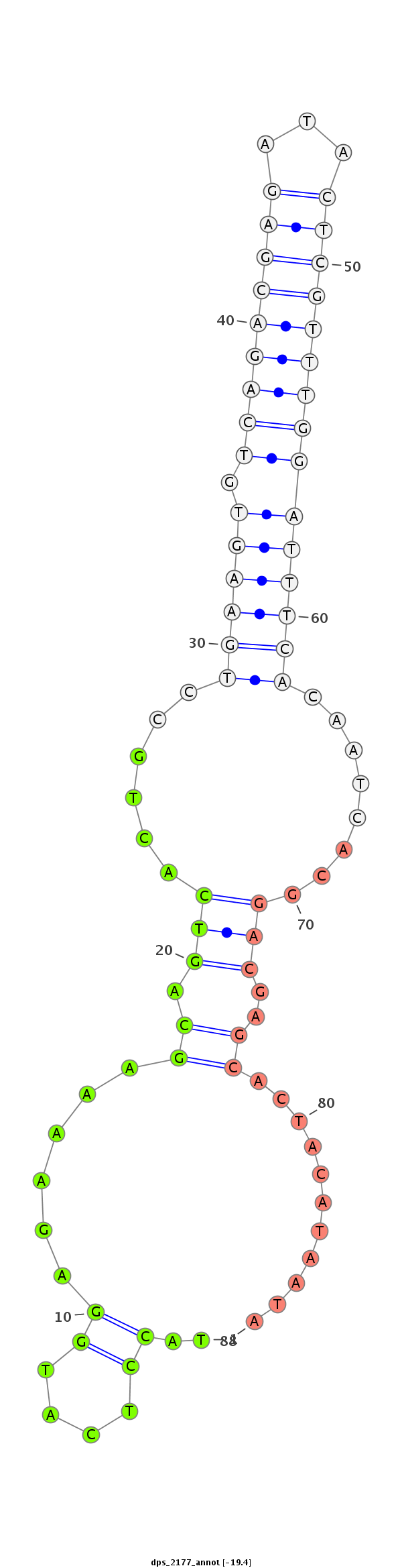
| .........................................................................................................................................CTGCTCGTGATGTATTATGT.............................................................................................. |
20 |
0 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................TCGTGATGTATTATGTGTGAT......................................................................................... |
21 |
0 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................GTGATGTGTAGATCAAATTG.......................................................................... |
20 |
0 |
5 |
0.80 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................GGATGATACATGAGCTTTTGT........................................................................................................................................................................................ |
21 |
0 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................GATGTGTAGATCAAATTGTAA....................................................................... |
21 |
0 |
5 |
0.60 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................GTGATGTGTAGATCAAATTGT......................................................................... |
21 |
0 |
5 |
0.60 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................GTCAGTGACGGACTTCACAGTCTGC.............................................................................................................................................. |
25 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................CTCGTGATGTATTATGTGTGA.......................................................................................... |
21 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................ATAGGATGATACATGAGCTT............................................................................................................................................................................................ |
20 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................ATAAAAACAAAAATAGGATGATACAT.................................................................................................................................................................................................. |
26 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................CTGCTCTTGATGTATTATGT.............................................................................................. |
20 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGATGTATTATGTGTGATG........................................................................................ |
21 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................CCTGCTCGTGATGTATTATG............................................................................................... |
20 |
0 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................AAAAACAAAAATAGGATGATACA................................................................................................................................................................................................... |
23 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GGTAGGTTCACCCGTGAAGC....................................... |
20 |
0 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................AATTGAAGAGGGGGGTAGGTT................................................... |
21 |
0 |
6 |
0.50 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................CAGTGACGGACTTCACAGTCT................................................................................................................................................ |
21 |
0 |
7 |
0.43 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................GTGTTAGTGCCTGCTCGTGAT....................................................................................................... |
21 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................GTGTTAGTGCCTGCTCGTGA........................................................................................................ |
20 |
0 |
5 |
0.40 |
2 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................GTGATGTATTATGTGTGATG........................................................................................ |
20 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................GATGTATTATGTGTGATGTGT..................................................................................... |
21 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................AGCTCCTGGAGTGGGATCTG.. |
20 |
0 |
6 |
0.33 |
2 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
| .........................................................GAGCTTTTGTATGGAGTACCT............................................................................................................................................................................. |
21 |
0 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................GTATTATGTGTGATGTGTA.................................................................................... |
19 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGGGTAGGTTCAACCGTCAGG........................................ |
21 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................................................CGGCAGTGACGGACTTCACAGTCT................................................................................................................................................ |
24 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................GGCAGGGACGGACTTCACAGTCTGCTC............................................................................................................................................ |
27 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................GTCAGTGACGGACTTCACAGTCTGGT............................................................................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................ATAGGATGATACATGAGCTTT........................................................................................................................................................................................... |
21 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................AAAAATAGGATGATACATGAGCT............................................................................................................................................................................................. |
23 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................GATGATACATGAGCTTTTGT........................................................................................................................................................................................ |
20 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................GTTAGTGCCTGCTCGTGATGT..................................................................................................... |
21 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................GTGTGATGTGTAGATCAAATT........................................................................... |
21 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................ATAGTGCCTGCTCGTGATGTAT................................................................................................... |
22 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................................................................................................GGCCGTGCACTAGCTCCTG.............. |
19 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................GGATGATACATGAGGTTTTGT........................................................................................................................................................................................ |
21 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................................CGTTGGCCGTGCACTAGCTCCT............... |
22 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................CCGTTGGCCGTGCACTAGCTCCT............... |
23 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................................GGTGTGATGGGTAGATCAAAT............................................................................ |
21 |
2 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................TGATGTGTAGATCAAATTG.......................................................................... |
19 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................GTTAGTGCCTGCTCGTGATG...................................................................................................... |
20 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................GTGTGATGTGTAGATCAAAT............................................................................ |
20 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................GTGTGATGTGTAGATCAAATTGT......................................................................... |
23 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................CTAGCTCCTGGAGTGGGAT..... |
19 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................GTAGATCAAGTTGTAAATTG................................................................... |
20 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................AATTGAAGAGGGGGGTAGGT.................................................... |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................GCTCTATGAGCAAACCTAAAGTGTT....................................................................................................................... |
25 |
0 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................................................................CCTGGAGTGGGATCTGCT |
18 |
0 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..............................................................................................................CTATGAGCAAACCTAAAGTGT........................................................................................................................ |
21 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................................................................TCCTGGAGTGGGATCTGCT |
19 |
0 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .................................................................................................................................................................................................................................CACTAGCTCCTGGAGTGGGAT..... |
21 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................AGCAAACCTAAAGTGTTAGTG................................................................................................................... |
21 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................AATGTGTGATGTGTAGATCAA.............................................................................. |
21 |
1 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................AGCTCCTGGAGTGGGATCTGC. |
21 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................GACGGACTTCACAGTCTGCTC............................................................................................................................................ |
21 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................GACTTCACAGTCTGCTCTAT......................................................................................................................................... |
20 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................................GTGAAGCACCATTGGCCGCGC......................... |
21 |
2 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................................................................CCGTGAAGCACCGTTGGCCGT........................... |
21 |
0 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................................................................CCGTGAAGCACCATTGGCCGC........................... |
21 |
2 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................AGCACCGTTGGCCGTGCACTA..................... |
21 |
0 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................TAGAGATGTATTATGTTTGAT......................................................................................... |
21 |
3 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ..................................................................................................................................................................GTATCGATCATATTGTAAATT.................................................................... |
21 |
3 |
11 |
0.09 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................GGGTAGGTTCACCCGTAACT........................................ |
20 |
3 |
12 |
0.08 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |