

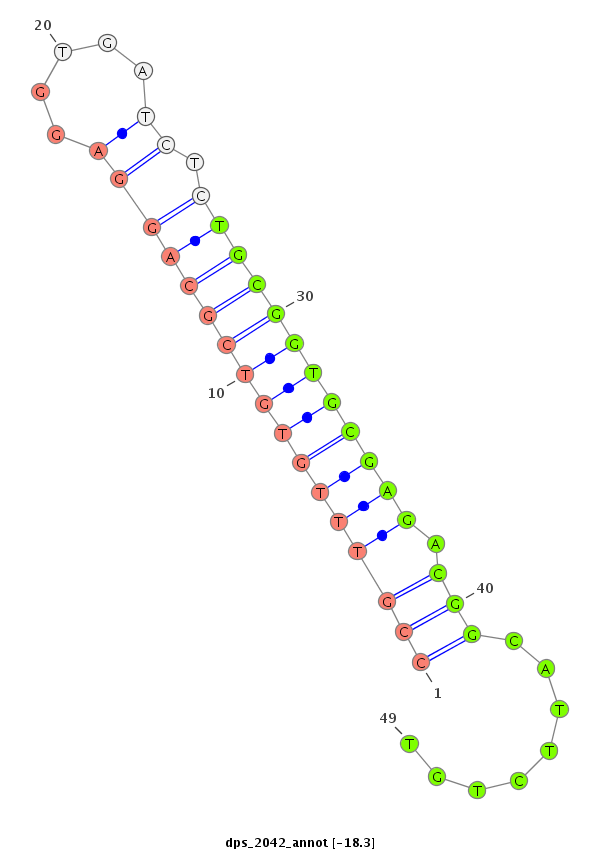
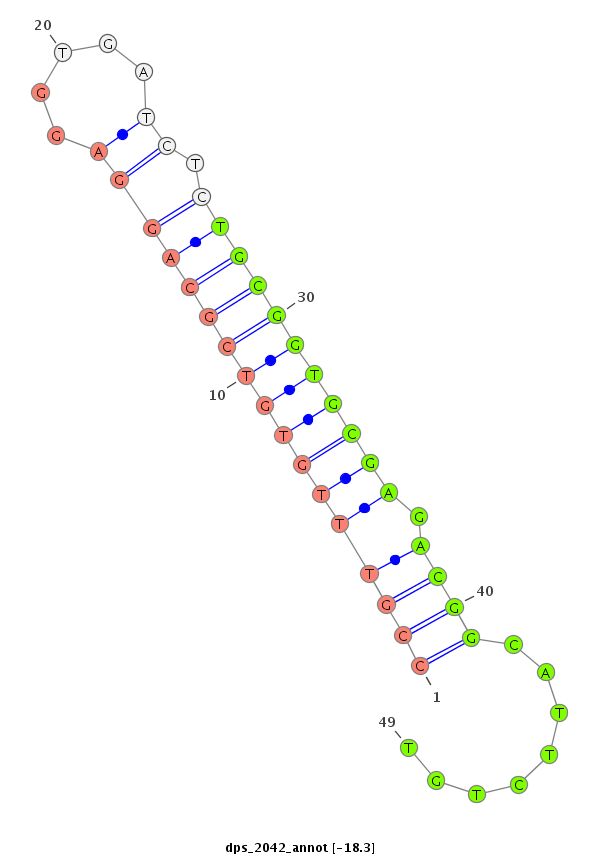
ID:dps_2042 |
Coordinate:Unknown_group_489:6579-6729 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
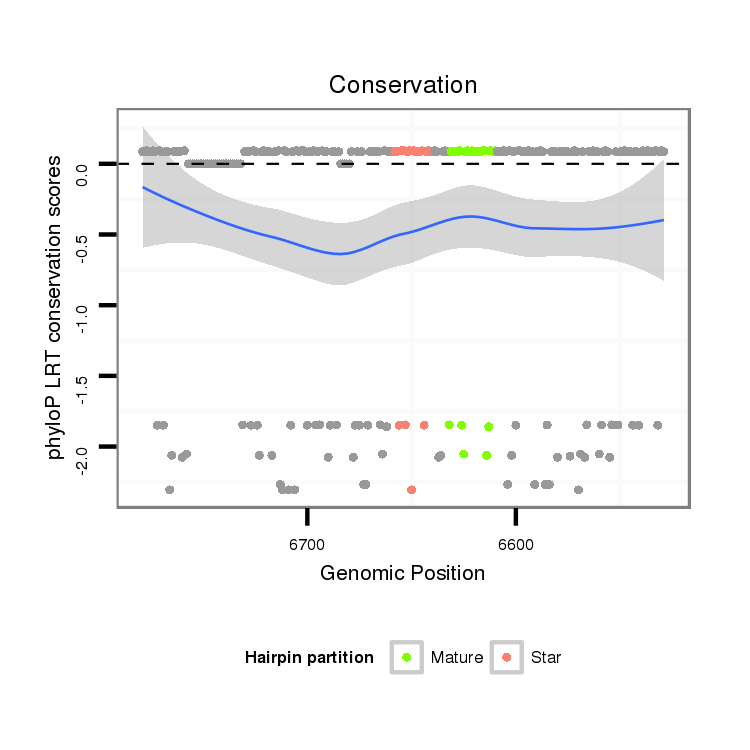
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.4 | -18.3 | -18.3 |
 |
 |
 |
CDS [Dpse\GA27969-cds]; exon [dpse_GLEANR_7209:3]; intron [Dpse\GA27969-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CGACGACGACGCGGTCGGCAACGCTTCCGACGAAGGCCGTGGATAAGGAGGGGTCGTCAGCCTGCACGAGGGGGAGATCGGCAACTTGGAGCGAGGAGAACAGCGGCCGGCCGCTCTCGGGCCGTTTGTGTCGCAGGAGGTGATCTCTGCGGTGCGAGACGGCATTCTGTGGCACCTTCGAACGCTGGCCCTCCACGCCAGCGCCGCAGGCGATCGAGCCAGAGATAGAGGAGGCGCGGCTGTGGGAGGAG *************************************************************************************************************************(((((((((((((((((.....)).))))))))).))))))........********************************************************************************* |
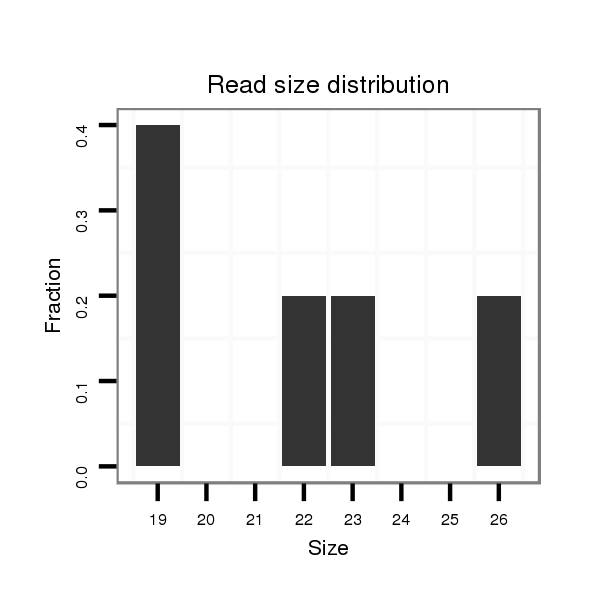
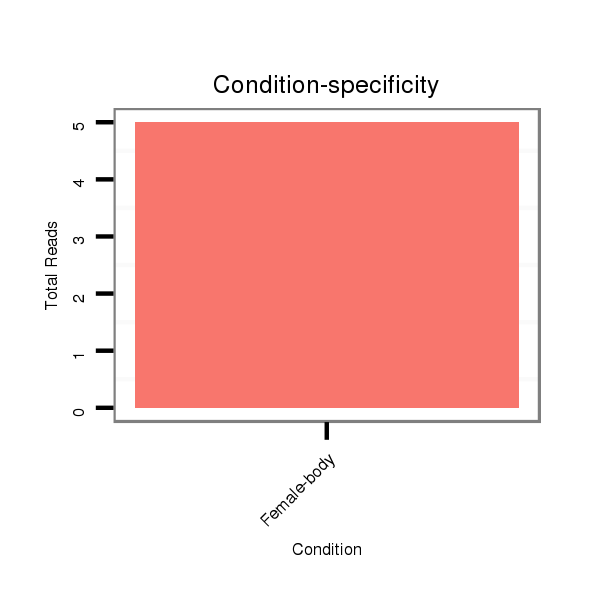
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
GSM343916 embryo |
M062 head |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................CAGGAGGTGATCTCTGCGGTGCGT.............................................................................................. | 24 | 1 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................TGCGGTGCGAGACGGCATTCTGT................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................TCAGCCTGCACGAGGGGGAGAA............................................................................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TGCGGTGCGTGACGGCATTCTGTGGCACC........................................................................... | 29 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................CAGGAGGTGATCTCTGCGG................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GTCGCAGGAGGTGATCTCTA...................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CCGTTTGTGTCGCAGGAGG............................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................CCTGCACGAGGGGGAGATCGGCAACT..................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CAGGAGGTGATCTCTGCGGTGC................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................TTCCGACGAAGGCCGTGGAT............................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................TCAGCCTGCACGAGGGGT................................................................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................TGATCTCTGCGGTGCGTGACGGCATTC.................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................CAGGAGGTGATCTCTGCGGTGCGTG............................................................................................. | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AGAGATACAGGAGGGGCGGCC.......... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......GACGCGGTCCACAACGCT.................................................................................................................................................................................................................................. | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TCGCAGGTAGTGATCTCTGGGG................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................ACCCTGCCGGAGGGGGAGATC............................................................................................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................CCAGCAGGAGGGGGAGATC............................................................................................................................................................................ | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGTGTCGCAGGAGGTAAA........................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................ATAGATGAGGCGGGGCTG......... | 18 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................ATGTGTCGCAGGAGGTGA............................................................................................................ | 18 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................GTTATCGGGCCGTTTGTGG........................................................................................................................ | 19 | 3 | 19 | 0.11 | 2 | 1 | 0 | 0 | 0 | 1 |
| .......................................................GTCAGGCTGGACGTGGGGGA................................................................................................................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................ACGAAGGCCGTGCATAAA............................................................................................................................................................................................................ | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................GGTGCGAGAGGTCATTTT................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GCTGCTGCTGCGCCAGCCGTTGCGAAGGCTGCTTCCGGCACCTATTCCTCCCCAGCAGTCGGACGTGCTCCCCCTCTAGCCGTTGAACCTCGCTCCTCTTGTCGCCGGCCGGCGAGAGCCCGGCAAACACAGCGTCCTCCACTAGAGACGCCACGCTCTGCCGTAAGACACCGTGGAAGCTTGCGACCGGGAGGTGCGGTCGCGGCGTCCGCTAGCTCGGTCTCTATCTCCTCCGCGCCGACACCCTCCTC
*********************************************************************************(((((((((((((((((.....)).))))))))).))))))........************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|
| ....................TGCGAAGGCTGCTTCCGGCACCTAT.............................................................................................................................................................................................................. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ....................................................................................................................................CGTCCTCCACTAGAGACGCCACGCT.............................................................................................. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ......................................................................................................................................................................................................GTCGCGGCGTCCGCCAGCTCGGT.............................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................AAGACACCGTGGAAGCTTGCGACCC.............................................................. | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................CCAGCAGTCGGACGTGCTCCC................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................GTCCTCCACTAGAGACGCCAC................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................CCGGCACCTATTCCTCCCCAGCAGT................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................GTCTCGGCGTCCGCTAGCTCGGTCT............................ | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................CCTCCGCGCTGACACCCT.... | 18 | 1 | 12 | 0.25 | 3 | 3 | 0 | 0 |
| ................................................................................................................................................GAGACGCGACGCTCTGCGCT....................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ...........................................................................CAAGCCGTTGAACATCACTC............................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ......................CGAAGGCTTCTTTCGGCATC................................................................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................TCTCTATCCCCTCTGTGCCG........... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 |
| .....................................................................................................................GCCCGGCAGAGACAGCGTA................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .............................TGAGTTCGGCACCTATTCCT.......................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | Unknown_group_489:6529-6779 - | dps_2042 | CG----ACGACGACGCGGTCGGCAACGCTTCCGACGAAGGCCGTGGATAAGGAGGGGTCGTCAGCCT--GCACGAGGGGGAG----------ATCGGCAAC-TTGGAGCGAGGAGAACAGCGGCCGGCCGCTCTCGGG-CCGTTTGTGTCGCAGGAGGTGATCTCTGCGGTGCGAGACGGCATTCTGTGGCACCTTCGAACGCT-----------GGCCCTCCACGCCAGCGCCGCAGGCGATCGAGCCAGAGATAGAGGAGGCGCGGCTGTGGGAGGAG |
| droPer2 | scaffold_6911:666-914 - | CGCCGGACGACAACACGCGCGGCCAA--------------------------GGGGATCAGCAGCCGCGGCAGCAGCAGCAGTATCAGCAGCATCAGCAGCACTGGCACGGG-----CCACAGGGAGCCGCCATTGGGCCCATTCGTCTCGCAAGAGGTGCGCTCCGCGGTAAGAGACGGCATGTTGTGGCACGTGCAAACGCTGCACATATCGTGGGCCTCGGGGCCCGCGCCTCAGCAGCCCGAGCAGGAGCCGGGGGAGGCACGACTGTGGGAAGAG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 02:39 AM