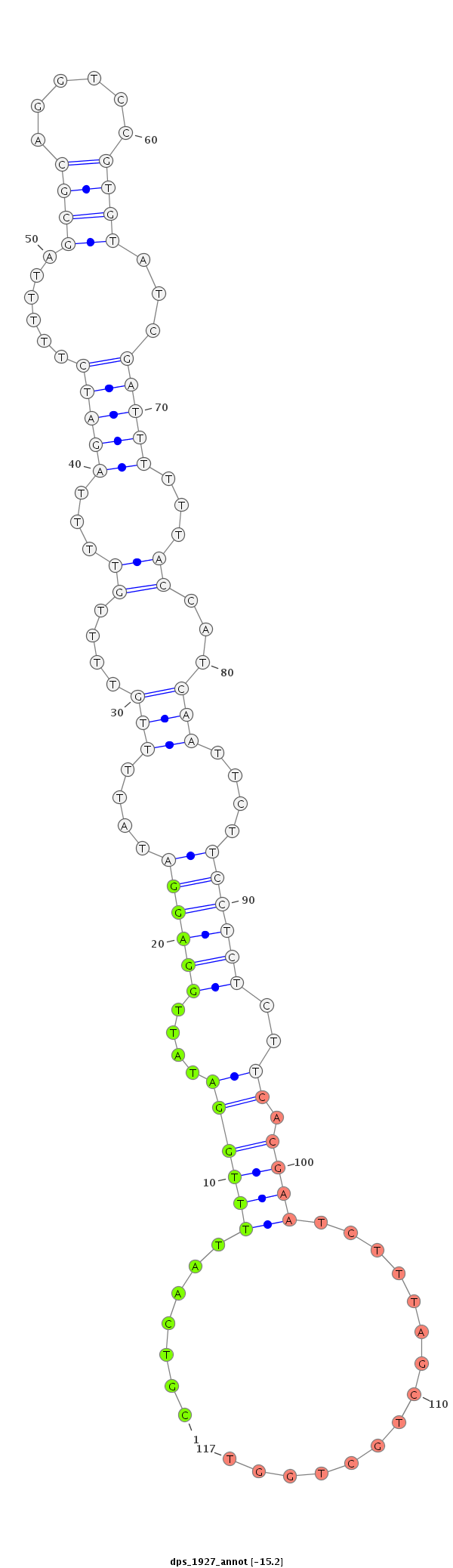
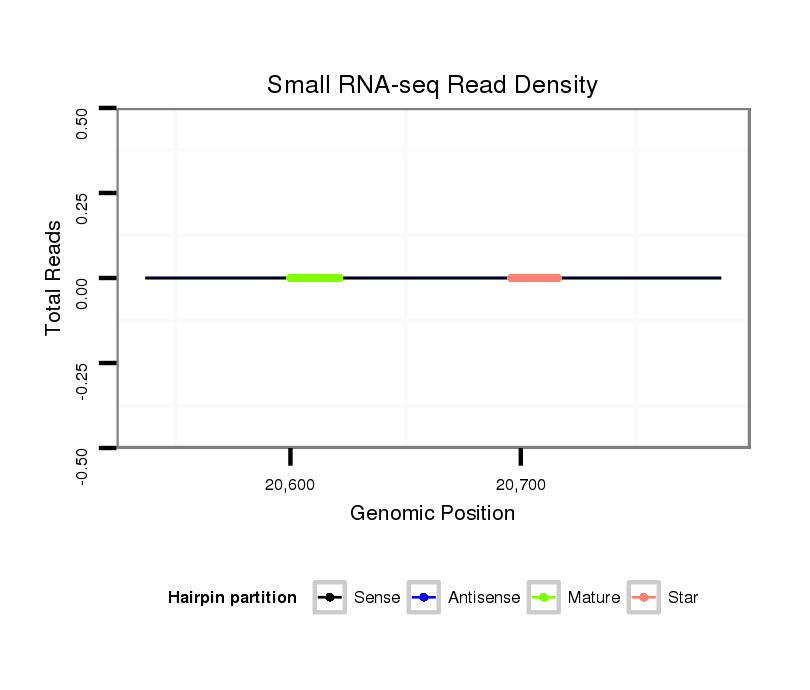

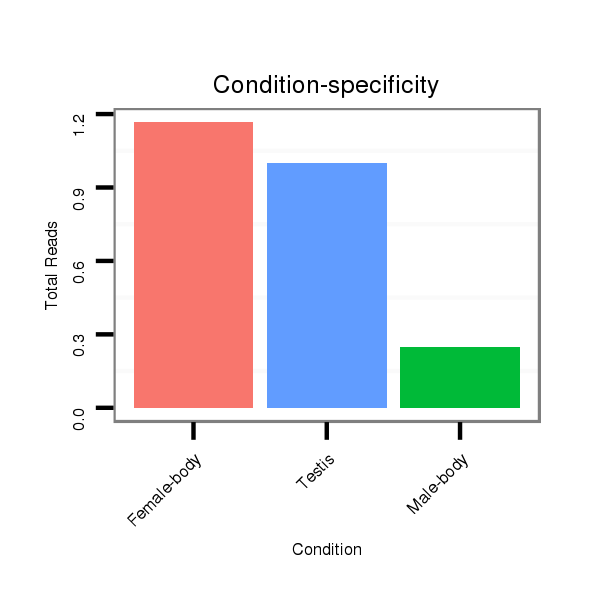
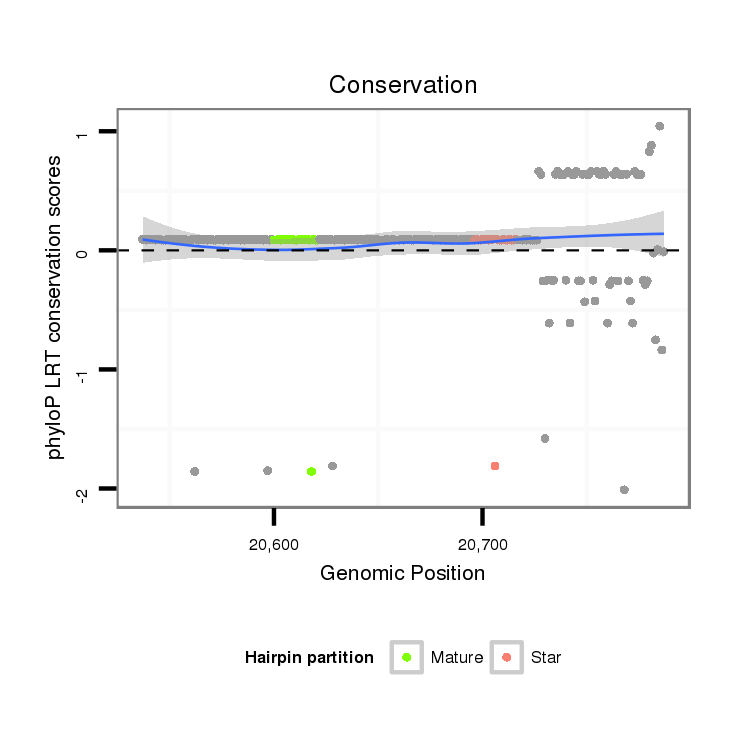
ID:dps_1927 |
Coordinate:Unknown_group_265:20587-20737 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -14.9 | -14.5 | -14.4 |
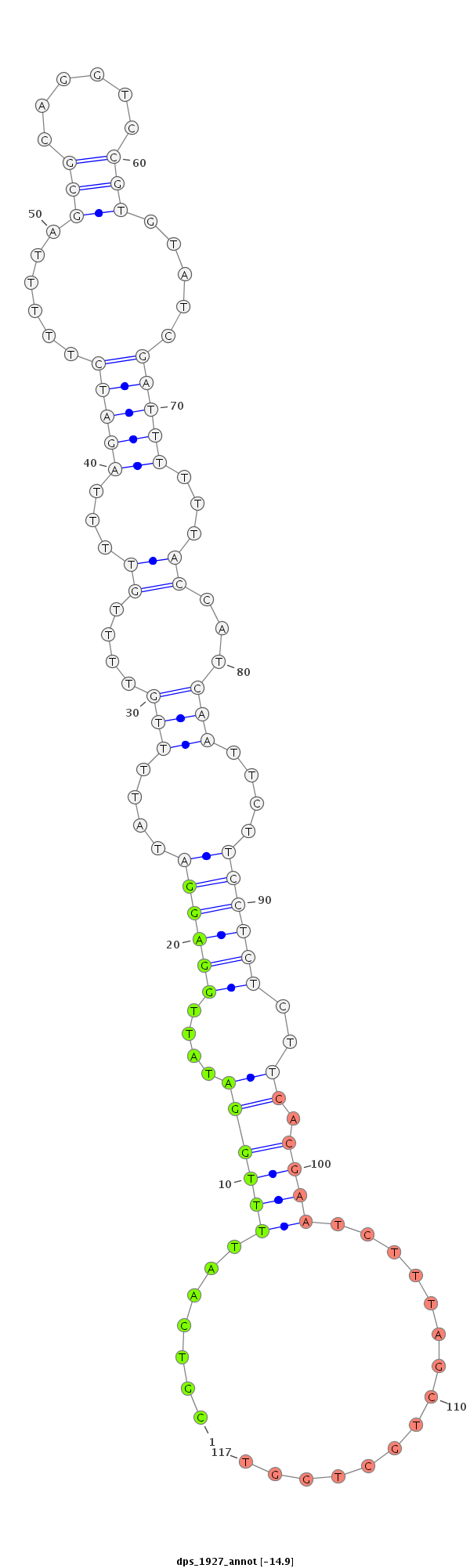 |
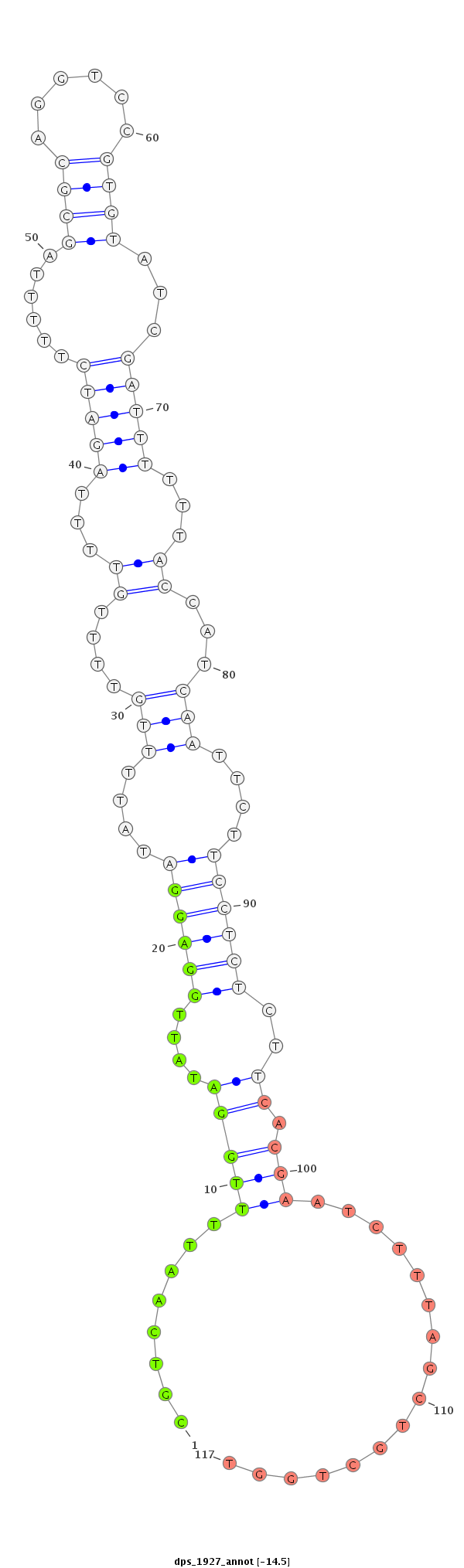 |
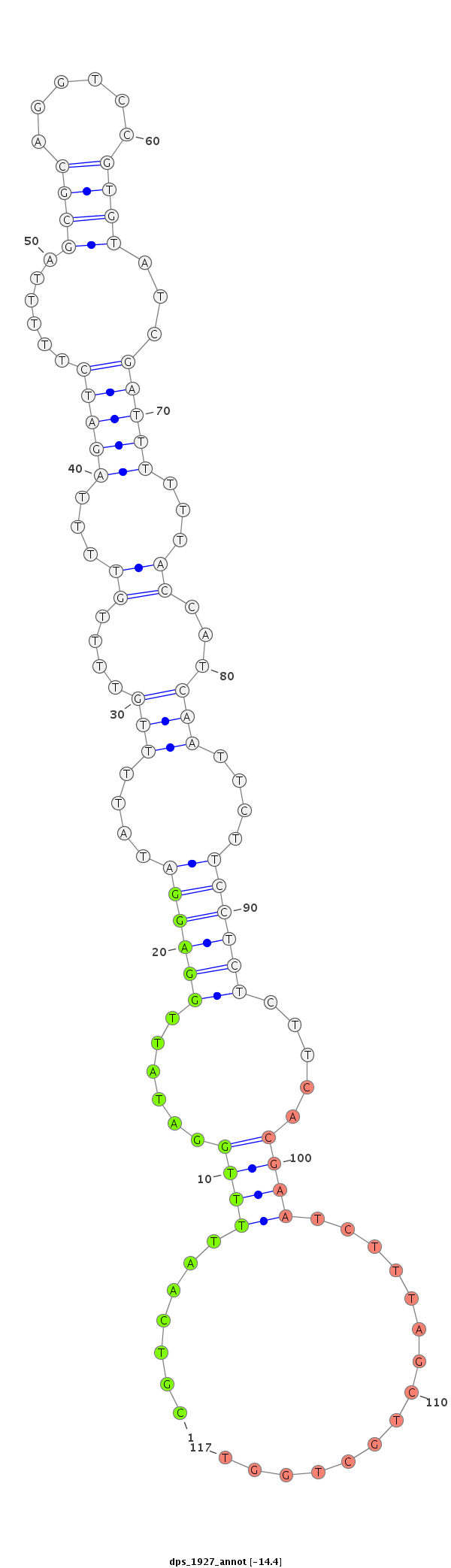 |
CDS [Dpse\GA28114-cds]; exon [dpse_GLEANR_7498:2]; intron [Dpse\GA28114-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTGATTGTTCCCATTTCATATGGGTGAGAATGAAGGTTTGACATTTAGTCACAAAATTCTACACGTCAATTTTGGATATTGGAGGATATTTTGTTTTGTTTTAGATCTTTTTAGCGCAGGTCCGTGTATCGATTTTTTACCATCAATTCTTCCTCTCTTCACGAATCTTTAGCTGCTGGTGCGTCTGCGGACGGAGAAGAGGCTACCCACCCCCCAAATGGAGGTCCTGCCGCCTCTGGCTTGCAGAAGGA ***************************************************************.......((((((....((((((....(((....((...(((((......((((......))))...)))))...))...)))....))))))..)).))))...............*********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
SRR902010 ovaries |
V112 male body |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................GACGGAGAAGAGGCTACCCA.......................................... | 20 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GACGGAGAAGAGGCTACCCAC......................................... | 21 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GACGGAGAAGAGGCTACCCACC........................................ | 22 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CGTCAATTTTGGATATTGGAGG...................................................................................................................................................................... | 22 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................CAATTTTGGATATTGGAGGAT.................................................................................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TGCGGACGGAGAAGAGGCTAACCA.......................................... | 24 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TCTGCGGACGGAGAAGAGGC................................................ | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................GCGGACGGAGAAGAGGCTACCCACC........................................ | 25 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CACGAATCTTTAGCTGCTGGT....................................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GAGGTCCTGCCGCCTCTGGCTTGCAGAAG.. | 29 | 0 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TTTTGGATATTGGAGGATATT................................................................................................................................................................. | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CGGACGGAGAAGAGGCTACCCAC......................................... | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................TTCTACACGTCAATTTTGGAT.............................................................................................................................................................................. | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................TGCCGCCTCTGGCTTGCAGAAGGA | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TTTTTAGCGCAGGTCCGTGTA........................................................................................................................... | 21 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................CAAAATTCTACACGTCAATTT................................................................................................................................................................................... | 21 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................TAGATCTTTTTAGCGCAGGTC................................................................................................................................. | 21 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................GATATTGGTGGGTATTTTGT............................................................................................................................................................. | 20 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AGAGGCTACACACCCACCA................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CCTGCCGCCTCTGGCTTGCA...... | 20 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................AAGAGGCTACACCCCCCAC.................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| ...................ATGGGTGAGGATTAAGCT...................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................GATCTTTTTAACGCAGC................................................................................................................................... | 17 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................TAGCTGCGGGTGCGTCCTC............................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
AACTAACAAGGGTAAAGTATACCCACTCTTACTTCCAAACTGTAAATCAGTGTTTTAAGATGTGCAGTTAAAACCTATAACCTCCTATAAAACAAAACAAAATCTAGAAAAATCGCGTCCAGGCACATAGCTAAAAAATGGTAGTTAAGAAGGAGAGAAGTGCTTAGAAATCGACGACCACGCAGACGCCTGCCTCTTCTCCGATGGGTGGGGGGTTTACCTCCAGGACGGCGGAGACCGAACGTCTTCCT
***********************************************************************.......((((((....((((((....(((....((...(((((......((((......))))...)))))...))...)))....))))))..)).))))...............*************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
|---|---|---|---|---|---|---|---|
| .............................................ATCAGTGTTTTAAGATGTGCA......................................................................................................................................................................................... | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 |
| ..............................................TCAGCGGGTTAAGATGTGCAGT....................................................................................................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 1 | 0 |
| ................................................................................................................................................TCAAGAATGAGAGAAGTG......................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 |
| .............AAAGTATACCGGCTCTTACGT......................................................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 |
| ..................................................................................................................GCGTCCAGGCACATAGCTAA..................................................................................................................... | 20 | 0 | 7 | 0.14 | 1 | 0 | 1 |
| ..................................................................................................................................CTTCAGAATGGTAGTTAAG...................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | Unknown_group_265:20537-20787 + | dps_1927 | TTGATTGTTCCCATTTCATATGGGTGAGAATGAAGGTTTGACATTTAGTCACAAAATTCTACACGTCAATTTTGGATATTGGAGGATATTTTGTTTTGTTTTAGATCTTTTTAGCGCAGGTCCGTGTATCGATTTTTTACCATCAATTCTTCCTCTCTTCACGAATCTTTAGCTGCTGGTGCGTCTGCGGACGGAGAAGAGGCTACCCACCCCCCAAAT--GGAGGTCCTGCCGCCTCTGGCTTGCAGAAGGA |
| droPer2 | scaffold_77:48376-48626 - | TTGATTGTTCCCATTTCATATGGGTAAGAATGAAGGTTTGACATTTAGTCACAAAATTCTGCACGTCAATTTTGGATATTGAAGGATATTTCGTTTTGTTTTAGATCTTTTTAGCGCAGGTCCGTGTATCGATTTTTTACCATCAATTCTTCCTCTCTTCACGAATCTTCAGCTGCTGGTGCGTCTGCGGACGAAGAAGAGGCTACCCACCCCCCAAAT--GGAGGTCCTGCCTCCTCTGGCTTGCAGAAGGA | |
| droBip1 | scf7180000396709:230403-230408 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGAGAA | |
| droEle1 | scf7180000491000:943547-943607 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAGGCGGGAGGCCAGCCATTCACCAGCTCCGGAGCATCTACCTCTGGTGGCCAACA--GGGC | |
| droEug1 | scf7180000408844:281595-281602 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACAGCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/18/2015 at 02:14 AM