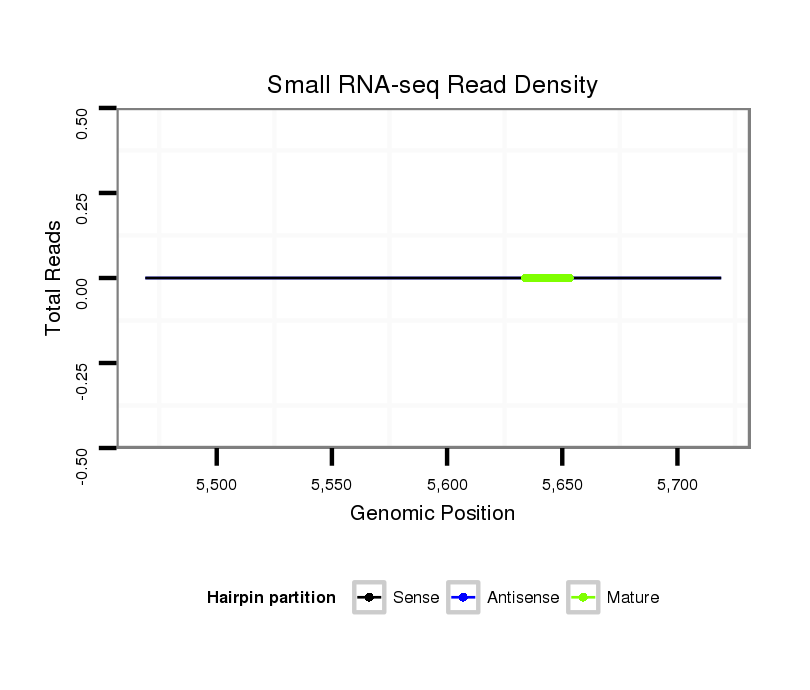
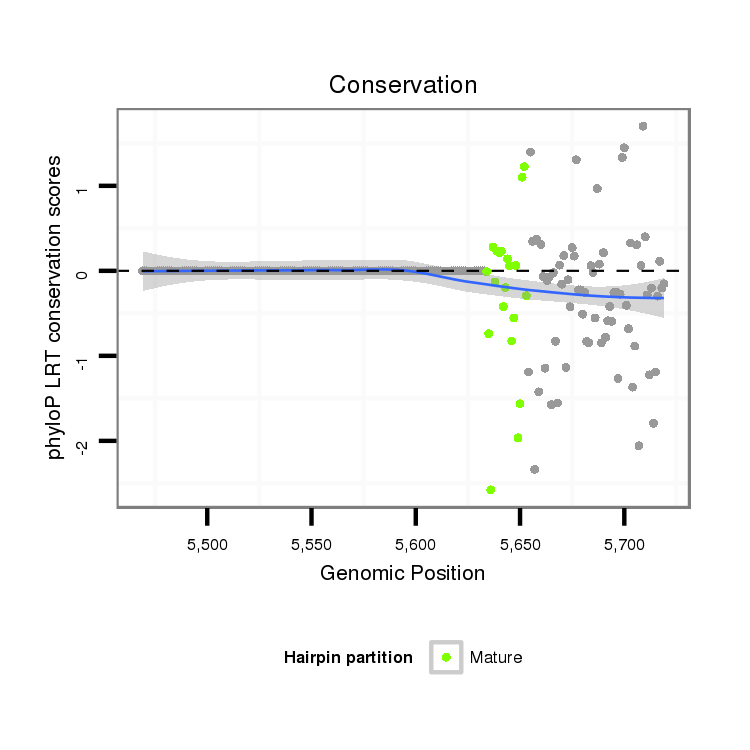
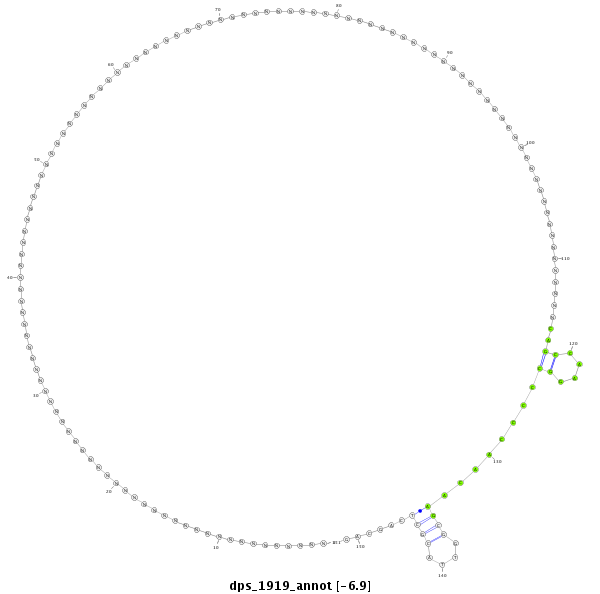
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------GCG--A-----AGCT--C---CC--CACTAAGGGACAACGTCTG----------GAAGCAAG-CTGCGCCGGT-------------------GTAGGG------GGATGCCAAGGCAC-CAACCCCCAAAACG-----GG | Size | Hit Count | Total Norm | Total | M044
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGCGCC...................................................................... | 23 | 1 | 35.00 | 35 | 2 | 33 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAAGGGACAACGTCTG----------GAAGCAAG-C............................................................................ | 25 | 1 | 13.00 | 13 | 1 | 11 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGCGC....................................................................... | 22 | 1 | 10.00 | 10 | 4 | 6 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CACTAAGGGACAACGTCTG----------GAAGCAAG-C............................................................................ | 28 | 1 | 9.00 | 9 | 3 | 6 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGCGCCGG.................................................................... | 25 | 1 | 7.00 | 7 | 6 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAACC................ | 24 | 1 | 7.00 | 7 | 1 | 6 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAACCC............... | 25 | 1 | 6.00 | 6 | 0 | 6 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAAC................. | 23 | 1 | 5.00 | 5 | 0 | 5 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGCGCCG..................................................................... | 24 | 1 | 4.00 | 4 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................T--C---CC--CACTAAGGGACAACGTCTG----------GAAG.................................................................................. | 27 | 1 | 4.00 | 4 | 0 | 4 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................T--C---CC--CACTAAGGGACAACGTCTG----------GAAGC................................................................................. | 28 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGTCTG----------GAAGCAAG-CTGCGCCGG.................................................................... | 24 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................AGGG------GGATGCCAAGGCAC-CAAC................. | 22 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................T-------------------GTAGGG------GGATGCCAAGGCAC-CA................... | 23 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................GCT--C---CC--CACTAAGGGACAACGTCTG----------GA.................................................................................... | 27 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................C---CC--CACTAAGGGACAACGTCTG----------GAAGC................................................................................. | 27 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAACA................ | 24 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................GCT--C---CC--CACTAAGGGACAACGTCTG----------G..................................................................................... | 26 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................T--C---CC--CACTAAGGGACAACGTCTG----------GAAGCA................................................................................ | 29 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGTCTG----------GAAGCAAG-CTGCGCC...................................................................... | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CC--CACTAAGGGACAACGTCTG----------GAAGC................................................................................. | 26 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGCGCCGGT-------------------G............................................... | 27 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................T--C---CC--CACTAAGGGACAACGTCTG----------G..................................................................................... | 24 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................T-------------------GTAGGG------GGATGCCAAGGCAC-..................... | 21 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................T-------------------GTAGGG------GGATGCCAAGGCAC-C.................... | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TG----------GAAGCAAG-CTGCGCCGGT-------------------GTAGG........................................... | 25 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAA.................. | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGCG........................................................................ | 21 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGCGCCGGT-------------------GTAGGG------GGATGCCAAG.......................... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CAACGTCTG----------GAAGCAAG-CTGCGCC...................................................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................T-------------------GTAGGG------GGATGCCAAGGCAC-CAAC................. | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACGTCTG----------GAAGCAAG-CTGC......................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................C--CACTAAGGGACAACGTCTG----------GAAGCAAG-............................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CACTAAGGGACAACGTCTG----------GAAGC................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................CT--C---CC--CACTAAGGGACAACGTCTG----------G..................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................AGGG------GGATGCCAAGGCAC-CAACC................ | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GAAGCAAG-CTGCGCCGGT-------------------GTAGG........................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGCGCCGGT-------------------GTAGGG------GGATGCCAAGG......................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGTCTG----------GAAGCAAG-CTGCGCCGGT-------------------................................................ | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGCGCCGGT-------------------GTAGGG------GGATGCC............................. | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................G-CTGCGCCGGT-------------------GTAGGG------GGATGCCA............................ | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................AG-CTGCGCCGGT-------------------GTAGGG------GGATG............................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................C---CC--CACTAAGGGACAACGTCTG----------GAAG.................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGCGCCGGT-------------------GTAGGG------GGATGCCT............................ | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................GACAACGTCTG----------GAAGCAAG-CTGCGCC...................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TG----------GAAGCAAG-CTGCGCCGGT-------------------GTAG............................................ | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGTCTG----------GAAGCAAG-CTGCGC....................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................T-------------------GTAGGG------GGATGCCAAGGCAC-CAACCCC.............. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAAGGGACAACGTCTG----------GAAGCAAC-............................................................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TCTG----------GAAGCAAG-CTGCGCCGGT-------------------GTA............................................. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................T-------------------GTAGGG------GGATGCCAAGGCAC-CAA.................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AGGGACAACGTCTG----------GAAGCAAG-CTGC......................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AGGGACAACGTCTG----------GAAGCAAG-CT........................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAACCCC.............. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................T--C---CC--CACTAAGGGACAACGTCTG----------GA.................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AAGCAAG-CTGCGCCGGT-------------------GTAGGG------GG.................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GCGCCGGT-------------------GTAGGG------GGATG............................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TAGGG------GGATGCCAAGGCAC-CAACCT............... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAAGGGACAACGTCTG----------GAAGCAAG-............................................................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................ACTAAGGGACAACGTCTG----------GAAGCAAG-C............................................................................ | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GCGCCGGT-------------------GTAGGG------GGATGCCAAG.......................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................GTAGGG------GGATGCCAAGGCAC-CA................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CAAG-CTGCGCCGGT-------------------GTAGGG------GG.................................. | 22 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................GCG--A-----AGCT--C---CC--CACTAAGGGAC........................................................................................................ | 22 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ...................................................................................................................................................................................T--C---CC--CACTAAGGGACAACGTC.................................................................................................. | 21 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TGCCAAGGCAC-CAACCCCCAAAACG-----G. | 26 | 18 | 0.17 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................GG------GGATGCCAAGGCAC-CAACCC............... | 22 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................GATGCCAAGGCAC-CAACCCCC............. | 21 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................GATGCCAAGGCAC-CAACCCCCAAA.......... | 24 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................AAGGCAC-CAACCCCCAAAACG-----G. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |