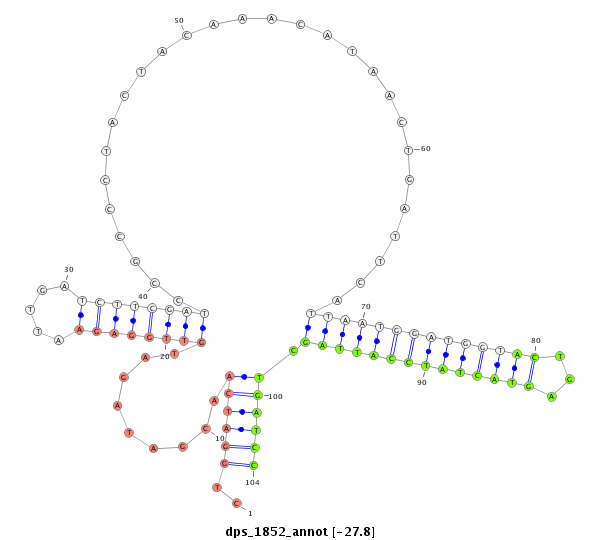
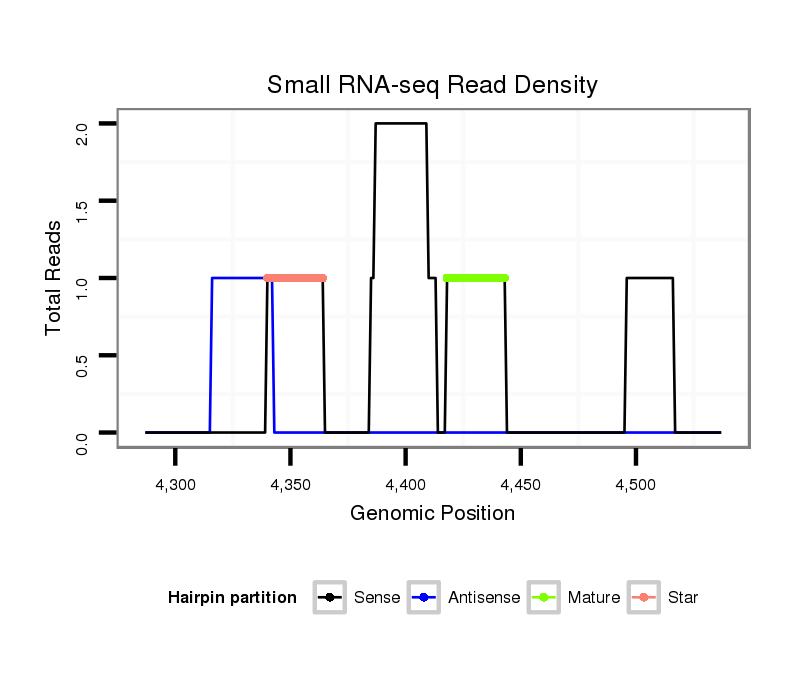
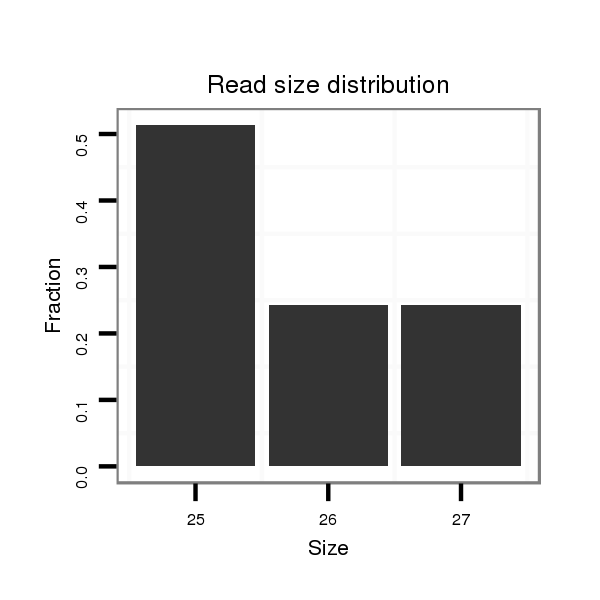
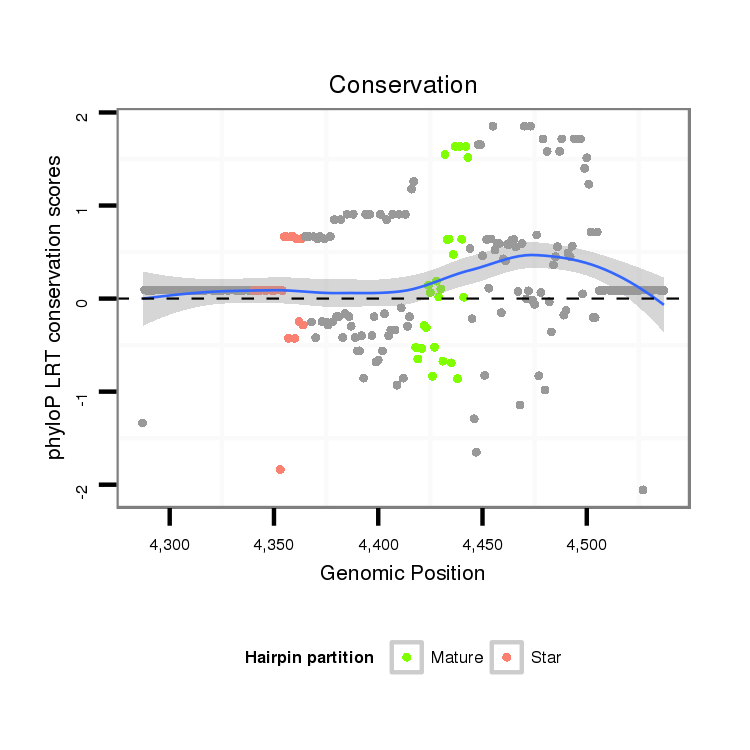
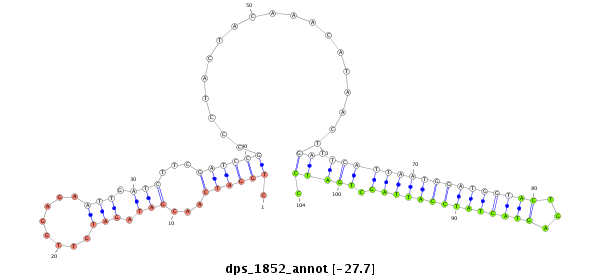
| G----------------------------------------------------------------------------------------------------------------------------------GTACCTGATAATGTA-------CAATTAGCTGATCCTCAATTCCTCAAGCCGCAACGTATTGATTTGTTGCTCGGTGCTGATATCTTTTTTGATATAT-------------------------------- | Size | Hit Count | Total Norm | Total | M046
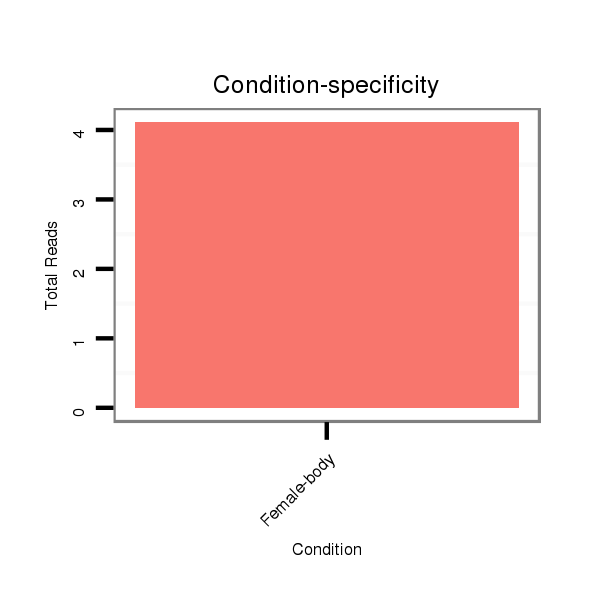
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| ............................................................................................................................................................................................TATTGATTTGTTGCTCGGTGC.................................................... | 21 | 20 | 0.40 | 8 | 6 | 0 | 0 | 0 | 2 | 0 |
| .........................................................................................................................................................................................ACGTATTGATTTGTTGCTCGG....................................................... | 21 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TGATTTGTTGCTCGGTGCTGA................................................. | 21 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................A-------CAATTAGCTGATCCTCAATT........................................................................................ | 21 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TATTGATTTGTTGCTCGGTGCTGAT................................................ | 25 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TA-------CAATTAGCTGATCCTCAAT......................................................................................... | 21 | 20 | 0.15 | 3 | 2 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................TACCTGATAATGTA-------CAATTAGCTGATCC.............................................................................................. | 28 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................ATTGATTTGTTGCTCGGTGCT................................................... | 21 | 20 | 0.10 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................AATGTA-------CAATTAGCTGATCCT............................................................................................. | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ..........................................................................................................................................ATAATGTA-------CAATTAGCTGATCC.............................................................................................. | 22 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................ATGTA-------CAATTAGCTGATCCTC............................................................................................ | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ............................................................................................................................................................................................TATTGATTTGTTGCTCGGTGCTG.................................................. | 23 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................GTACCTGATAATGTA-------CAATTAGCT................................................................................................... | 24 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TGATTTGTTGCTCGGTGCTG.................................................. | 20 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GTA-------CAATTAGCTGATCCTCAA.......................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CCTGATAATGTA-------CAATTAGCTGATCC.............................................................................................. | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TGATTTGTTGCTCGGTGCTGATAA.............................................. | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTGATAATGTA-------CAATTAGCT................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................GTACCTGATAATGTA-------CAATTAGCTGATC............................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................ATTTGTTGCTCGGTGCTGA................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................TATTGATTTGTTGCTCGGTGCTGA................................................. | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................CAAGCCGCAACGTATTGATT................................................................. | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CGTATTGATTTGTTGCTCGGTGCTGAT................................................ | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................TTGATTTGTTGCTCGGTGCTGT................................................. | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................GTACCTGATAATGTA-------CAATTAGCTG.................................................................................................. | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................TAATGTA-------CAATTAGCTGATCCTCAAT......................................................................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................GTATTGATTTGTTGCTCGGTGCTGATA............................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................TTGATTTGTTGCTCGGTGCTGATA............................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................TGTTGCTCGGTGCTGATATCTTTTTTG...................................... | 27 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGTGCTGATATCTTTTTTGAA.................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TGTA-------CAATTAGCTGATCCTCAA.......................................................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CCTGATAATGTA-------CAATTAGCTGATCCA............................................................................................. | 27 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATGTA-------CAATTAGCTGATCCTCAATTC....................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................ATTTGTTGCTCGGTGCTGATA............................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................ATAATGTA-------CAATTAGCTGATC............................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................CCTGATAATGTA-------CAATTAGCTGATCCTC............................................................................................ | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GTA-------CAATTAGCTGATCCTCAATTCC...................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................ATTGATTTGTTGCTCGGTGCTTT................................................. | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................TGTA-------CAATTAGCTGATCCTCAATT........................................................................................ | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................ACCTGATAATGTA-------CAATTAG..................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATGTA-------CAATTAGCTGATCCTCAATT........................................................................................ | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................GTACCTGATAATGTA-------CAATTAGCTGAAA............................................................................................... | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TTGATTTGTTGCTCGGTGCTGAT................................................ | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................................................AACGTATTGATTTGTTGCTCGGTGT.................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................GTATTGATTTGTTGCTCGGTGCTGATATC............................................. | 29 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CGTATTGATTTGTTGCTCGGT...................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................AATTAGCTGATCCTCAATT........................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................TACCTGATAATGTA-------CAATTAGCTG.................................................................................................. | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TTGTTGCTCGGTGCTGATAT.............................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................TGTA-------CAATTAGCTGATCCTCAAT......................................................................................... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GATTTGTTGCTCGGTGCTG.................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CCTGATAATGTA-------CAATTAGCT................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TGCTCGGTGCTGATATCTTTTTTGAT.................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ACGTATTGATTTGTTGCTCGGT...................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................GATTTGTTGCTCGGTGCTGAT................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................AACGTATTGATTTGTTGCTCGG....................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATGTA-------CAATTAGCTGATCC.............................................................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................ATTGATTTGTTGCTCGGTGCTGATC............................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................ATTTGTTGCTCGGTGCTGT................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |