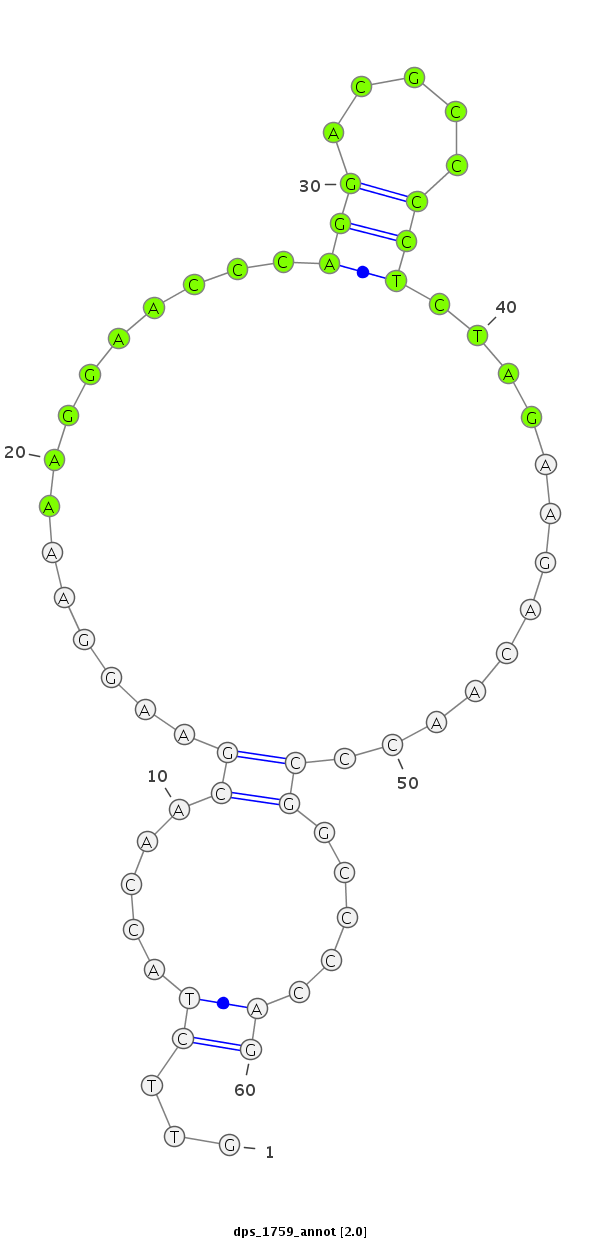
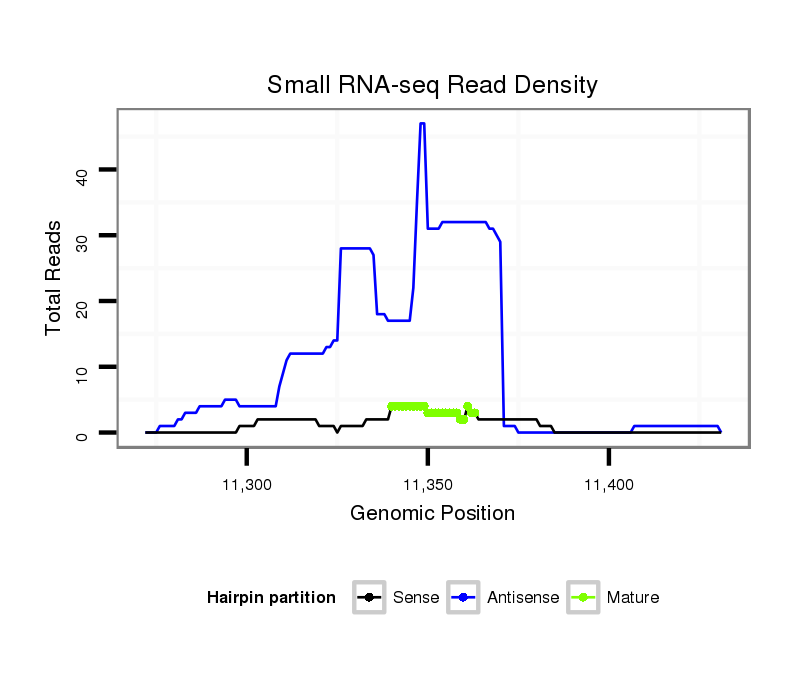
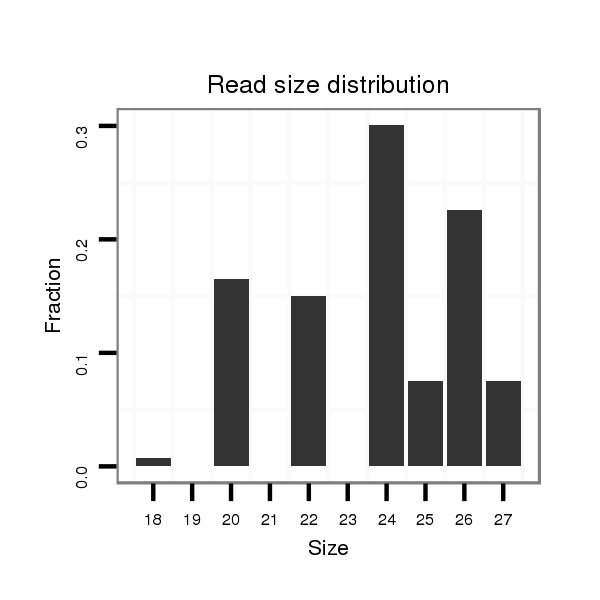
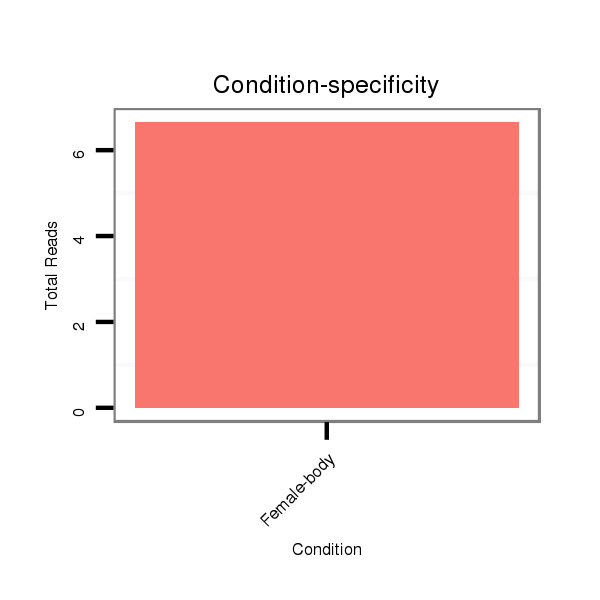
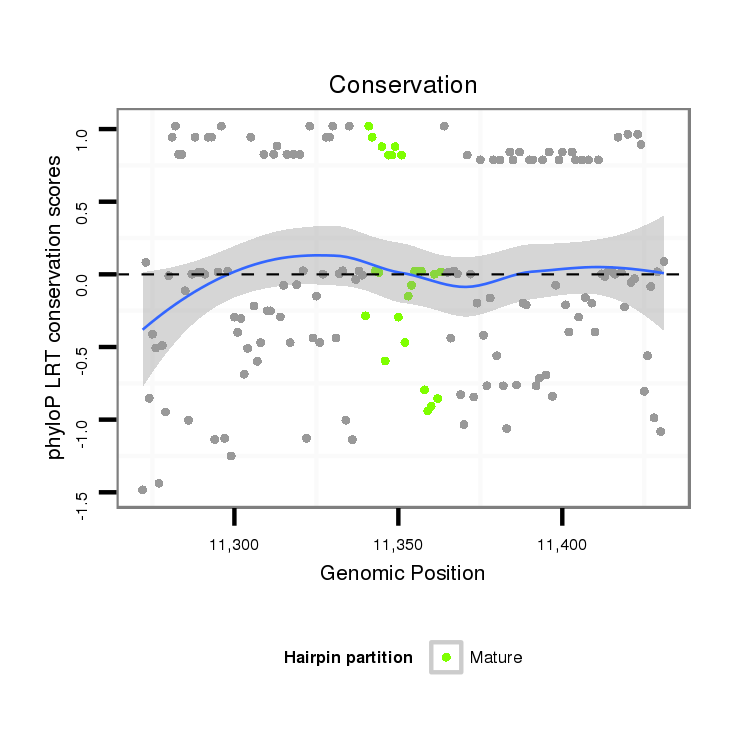
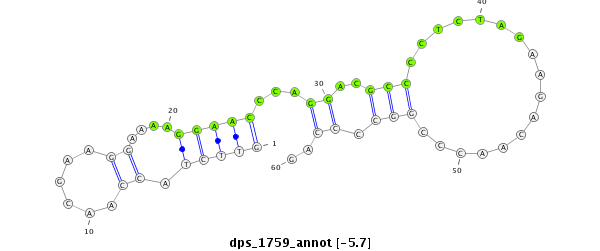
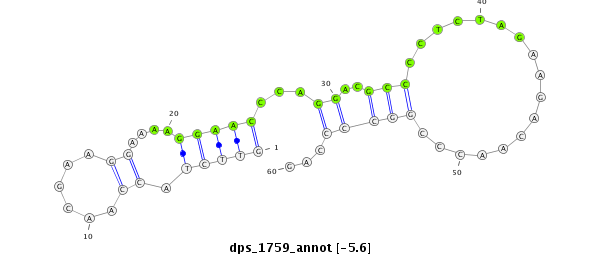
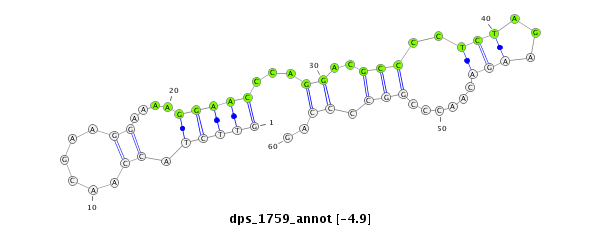
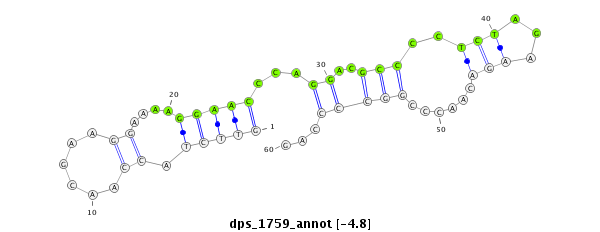
ID:dps_1759 |
Coordinate:Unknown_group_12:11322-11381 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -5.7 | -5.6 | -4.9 | -4.8 |
 |
 |
 |
 |
CDS [Dpse\GA23217-cds]; CDS [Dpse\GA23217-cds]; exon [dpse_GLEANR_11789:3]; exon [dpse_GLEANR_11789:2]; intron [Dpse\GA23217-in]
| Name | Class | Family | Strand |
| Gypsy9-I_Dpse-int | LTR | Gypsy | + |
| ##################################################------------------------------------------------------------################################################## TAAGCAAAGCAGCGAAGAAAGCGATCCGGTCTCCCGAGGGCACTGACGCGGTTCTACCAACGAAGGAAAAGGAACCCAGGACGCCCCTCTAGAAGACAACCCGGCCCCAGCAACAACGGCCGCCACGCAGCAGCCAGGCGCCCGACGTACCACCTCCGAC **************************************************...((.....((...............(((.....))).............)).....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
|---|---|---|---|---|---|---|
| ....................................................................AAGGAACCCAGGACGCCCCTCTAG.................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 |
| ..........................CGGTCTCCCGAGGGCACTGACG................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 |
| .............................................................GAAGGAAAAGGAACCCAGGACGCCCC......................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................TACCAACGAAGGAAAAGGAACCCA.................................................................................. | 24 | 0 | 2 | 1.00 | 2 | 2 |
| ...............................TCCCGAGGGCACTGACGCGGTT........................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TAGAAGACAACCCGGCCCCA................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TAGAAGACAACCCGGCCCCAGCAA............................................... | 24 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................AAGGAACCCAGGACGCCCCTCT...................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................TCTACCAACGAAGGAAAAGGAACCCA.................................................................................. | 26 | 0 | 2 | 0.50 | 1 | 1 |
| ......................................................TACCAACGAAGGAAAAGGAACCCAGGA............................................................................... | 27 | 0 | 2 | 0.50 | 1 | 1 |
| ....................................AGGGCACTGACGCGGTTCTACCAACG.................................................................................................. | 26 | 0 | 2 | 0.50 | 1 | 1 |
| .....................................................CTACCAACGAAGGAAAAGGAACCCA.................................................................................. | 25 | 0 | 2 | 0.50 | 1 | 1 |
| .......................................................ACCCAAGAAGGAAAAGGAAC..................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 1 |
| ....................................................................AAGGAACCCAGGACGCCCCT........................................................................ | 20 | 0 | 20 | 0.10 | 2 | 2 |
| ..........................................................................................AGAAGACAACCCGGCCCCAGCA................................................ | 22 | 0 | 17 | 0.06 | 1 | 1 |
| ..................................................TTTCTACCAGCGAAGCAAAA.......................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 1 |
| ..........................................................................................AGAAGACAACCCGGCCCCAGC................................................. | 21 | 0 | 17 | 0.06 | 1 | 1 |
| ..........................................................................................................................CCACGCAGCAGCCAGGCGC................... | 19 | 0 | 19 | 0.05 | 1 | 1 |
| ....................................................................AAGGAACCCAGGACGCCC.......................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 |
|
ATTCGTTTCGTCGCTTCTTTCGCTAGGCCAGAGGGCTCCCGTGACTGCGCCAAGATGGTTGCTTCCTTTTCCTTGGGTCCTGCGGGGAGATCTTCTGTTGGGCCGGGGTCGTTGTTGCCGGCGGTGCGTCGTCGGTCCGCGGGCTGCATGGTGGAGGCTG
**************************************************...((.....((...............(((.....))).............)).....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
M059 embryo |
V112 male body |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................ATGGTTGCTTCCTTTTCCTTGGGT.................................................................................. | 24 | 0 | 2 | 14.00 | 28 | 27 | 1 | 0 | 0 | 0 |
| ...........................................................................GGTCCTGCGGGGAGATCTTCTGTT............................................................. | 24 | 0 | 1 | 11.00 | 11 | 11 | 0 | 0 | 0 | 0 |
| ............................................................................GTCCTGCGGGGAGATCTTCTGTT............................................................. | 23 | 0 | 1 | 11.00 | 11 | 11 | 0 | 0 | 0 | 0 |
| ..........................................................................GGGTCCTGCGGGGAGATCTTCTGTT............................................................. | 25 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 |
| .....................................CCCGTGACTGCGCCAAGATGGTTGCTT................................................................................................ | 27 | 0 | 2 | 3.50 | 7 | 6 | 0 | 1 | 0 | 0 |
| ......................................CCGTGACTGCGCCAAGATGGTTGCTT................................................................................................ | 26 | 0 | 2 | 2.50 | 5 | 5 | 0 | 0 | 0 | 0 |
| .......................................CGTGACTGCGCCAAGATGGTTGCTT................................................................................................ | 25 | 0 | 2 | 2.50 | 5 | 5 | 0 | 0 | 0 | 0 |
| ..............................................GCGCCAAGATGGTTGCTTCCTTTTCCT....................................................................................... | 27 | 0 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................................GTGACTGCGCCAAGATGGTTGCTT................................................................................................ | 24 | 0 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...............TCTTTCGCTAGGCCAGAGGGCTCCCGT...................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................GTCCTGCGGGGAGATCTTCTGT.............................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................CTAGGCCAGAGGGCTCCCGTGACT.................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CAAGATGGTTGCTTCCTTTTCCTTGGGT.................................................................................. | 28 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................GGTCCTGCGGGGAGATCTTC................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................AGATGGTTGCTTCCTTTTCCTTGGGT.................................................................................. | 26 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................GACTGCGCCAAGATGGTTGCTTCCT............................................................................................. | 25 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................AATAGGCCAGAGGGCTCCCGTGACT.................................................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TCCGCGGGCTGCATGGTGGAGGCT. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....GTTTCGTCGCTTCTTTCGCTAG...................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................CGGGGAGATCTTCTGTTGGGC......................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........CGCTTCTTTCGCTAGGCCAGAGGGCT........................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........GTCGCTTCTTTCGCTAGGCCAGAGGGC............................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................CCCGTGACTGCGCCAAGATGGTTGCT................................................................................................. | 26 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................GGTCCTGCGGGGAGATCTTCTG............................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................TCCCGTGACTGCGCCAAGATGGTTGCTT................................................................................................ | 28 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................TGGGTCCTGCGGGGAGATCTTCTGTT............................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................CCAAGATGGTTGCTT................................................................................................ | 15 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................GGTTGCTTCCTTTTCCTTGGGT.................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................GGATCGTCTGTTGGGCAGGGG.................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................CGTGACTGCGCCAAGATGGTTGCT................................................................................................. | 24 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................GACTGCGCCAAGATGGTTGCTTCCTT............................................................................................ | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCCAAGATGGTTGCTTCCTTTTCCT....................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................GGGTCCCGGGACTACGCCAA........................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................GGTGCCGGCCGTGCGTCG............................. | 18 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................ATGGTTGCTTCCCTTTCCTTGGGT.................................................................................. | 24 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | Unknown_group_12:11272-11431 + | dps_1759 | TAAGCAA--AGCAGCGAAGAA-------AGCGATCCGGTCTCCC-----------GAGGGCACTGACGCGGTTCTAC-----CAACGAA---GGAAAAGGAA--------------CCCAGGACGCCCCTCTAGAAGACAACCC--GGCCCCAGCAACAACGGCCGCCACGCAGCAGC-----------CAGGCGCCCGACGT-----ACCACCTC-----CGAC |
| droPer2 | scaffold_55:34611-34770 + | TAAGCAA--AGCAGCGAAGAA-------AGCGATCCGGTCTCCC-----------GAGGGCACTGACGCGGTTCTAC-----CAACGAA---GGAAAAGGAA--------------CCCAGGACGCCCCTCTAGAAGACAACCC--GGCCCCAGCAACAACGGCCGCCACGCAGCAGC-----------CAGGCGCCCGACGT-----ACCACCTC-----CGAC | |
| droKik1 | scf7180000299893:23226-23366 + | CGCGTGG--ACCAGCGAAGGG-------AGCCATGCTCTAACCCACGGGACCAGCCCG--CAGAGCCACGCTCTGAC-----CAGCGAAAAACGACAAGGGA--------------GCCACGCTCCCCACCTGAAGAACAGCCA-------------------------TC------------------------CACGACAC-----ACCACTCAGAGCCAAGC | |
| droFic1 | scf7180000452674:140-294 - | CAAACCC--GGCAGCAGAGAACCCTCAAAGCCATCCC-----CAGCCGCAGCACCGAGAACAGAGACACAGTC--AC-----CAGAAGAACACGACCAGAA-----------------------GCCCA-GTCGAAGGATA-CC--AGCAACCGAGACCATGGCAGCGACGCAGAAGGAAGA-------CCGAAGACCAACG----------------------T | |
| droEle1 | scf7180000488233:21944-21969 + | AAAGCAC--AG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAACACCGAGGAAAGC | |
| droRho1 | scf7180000776997:11435-11616 + | GAGAAGCGGCGCA---CCGAA-------CGCGGTGTGCGCGGCA-------------------------GCTTCTCCTGATACAACGCA---GCTCCAGGAAAACCCCGTGGCAGCCCCACGACGTTTGGTGGGAAAACCCCATAGGGATACAGTCACTACAGCTCCCAAGCAACAGGAACGTGTACGACCGGCGCCTGCCGTGGATAACCACTTC-----CACC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/18/2015 at 01:24 AM