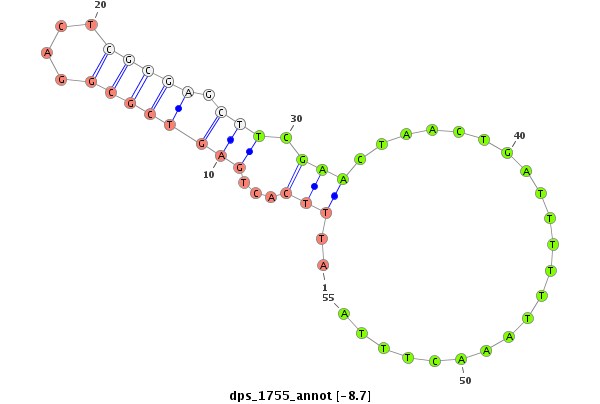
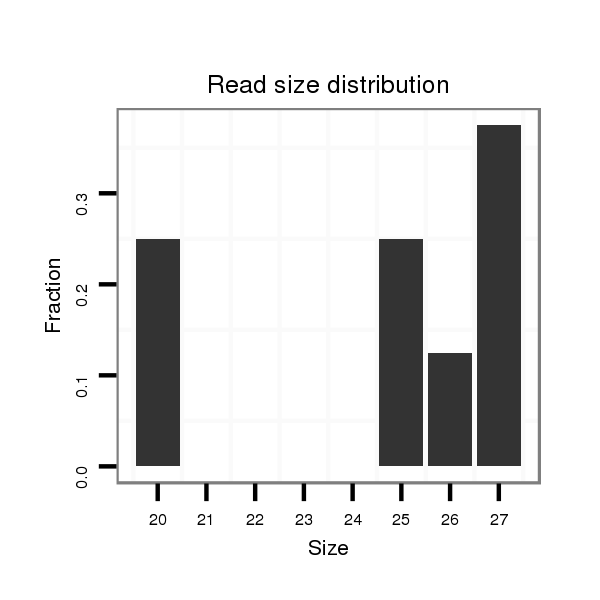
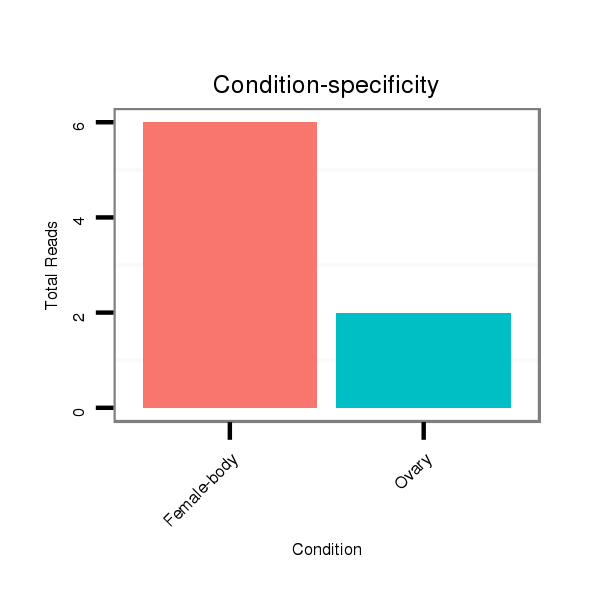
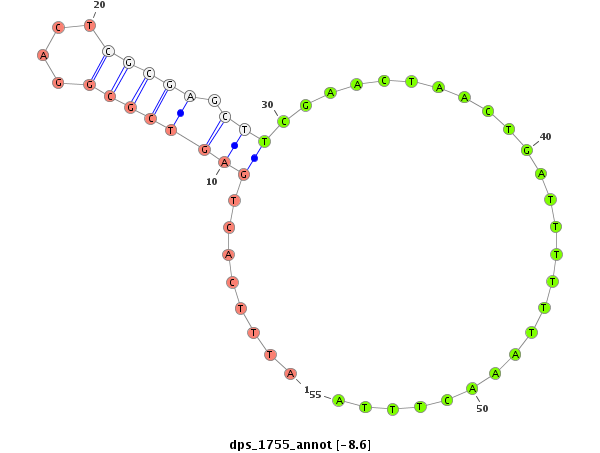
| TATTGCGTAGAAGCGAAAGATTACCGTTCCACCAGGGTGGCCTGGTCCTCTTCCTGGCTCTAGATGTCGGGCATGCTGCGCGCTGCGCATCTAATAATTCCCTGGAGAGGGTCTCTAGAGACTTTTCTATGTCTTCAGCTGATTTTACTAGGCCCGGGCCAGTCGCGAGTTGTGTCGAGATTGCCTC--------------------------T-------------------------------------------TCCCCCACGTTAGCTATGCGTATGTTAGTTGGTAGTAAA | Size | Hit Count | Total Norm | Total | M026
Head | M043
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| ...............................................................................................................................TATGTCTTCAGCTGATTTTACTAGGC............................................................................................................................................... | 26 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TATGTCTTCAGCTGATTTTACTAGGCC.............................................................................................................................................. | 27 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................ATGTCTTCAGCTGATTTTACTAGGCC.............................................................................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCTAGATGTCGGGCATGCTGCGCGC..................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................TCCCCCACGTTAGCTATGCGTATGT.............. | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TAGATGTCGGGCATGCTGCGCG...................................................................................................................................................................................................................... | 22 | 6 | 1.00 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| .......................................................................................................................GACTTTTCTATGTCTTCAGCTGATTT....................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TCGAGATTGCCTC--------------------------T-------------------------------------------TCCCCCACGTT............................ | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TAGAAGCGAAAGATTACCGTTCCACC....................................................................................................................................................................................................................................................................... | 26 | 3 | 1.00 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCTAATAATTCCCTGGAGAGGGTCTC..................................................................................................................................................................................... | 26 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCTAATAATTCCCTGGAGAGGGTC....................................................................................................................................................................................... | 24 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........GAAGCGAAAGATTACCGTTCCACCA...................................................................................................................................................................................................................................................................... | 25 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TGCGCATCTAATAATTCCCTGGAGAGG.......................................................................................................................................................................................... | 27 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCTAATAATTCCCTGGAGAGGGT........................................................................................................................................................................................ | 23 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TTTTACTAGGCCCGGGCCAGTCGCGAG............................................................................................................................... | 27 | 7 | 0.29 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TCCTGGCTCTAGATGTCGGGCATGCTG.......................................................................................................................................................................................................................... | 27 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................TACCGTTCCACCAGGGTGGC............................................................................................................................................................................................................................................................... | 20 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TATTGCGTAGAAGCGAAAGATTACC............................................................................................................................................................................................................................................................................... | 25 | 9 | 0.22 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TGGCTCTAGATGTCGGGCATGCTGCG........................................................................................................................................................................................................................ | 26 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TATTGCGTAGAAGCGAAAGATTACCGT............................................................................................................................................................................................................................................................................. | 27 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................GGCTCTAGATGTCGGGCATGCTGCA........................................................................................................................................................................................................................ | 25 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...TGCGTAGAAGCGAAAGATTACCGTTC........................................................................................................................................................................................................................................................................... | 26 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TTTACTAGGCCCGGGCCAGTCGCGAG............................................................................................................................... | 26 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TAGATGTCGGGCATGCTGCGC....................................................................................................................................................................................................................... | 21 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TCCTCTTCCTGGCTCTAGATGTCG................................................................................................................................................................................................................................... | 24 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TTTTACTAGGCCCGGGCCAGTCGCGA................................................................................................................................ | 26 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TGGTCCTCTTCCTGGCTCTAGATGTC.................................................................................................................................................................................................................................... | 26 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TTCCACCAGGGTGGCCTGGTCCTCT..................................................................................................................................................................................................................................................... | 25 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TCCACCAGGGTGGCCTGGTCCTCTTC................................................................................................................................................................................................................................................... | 26 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |