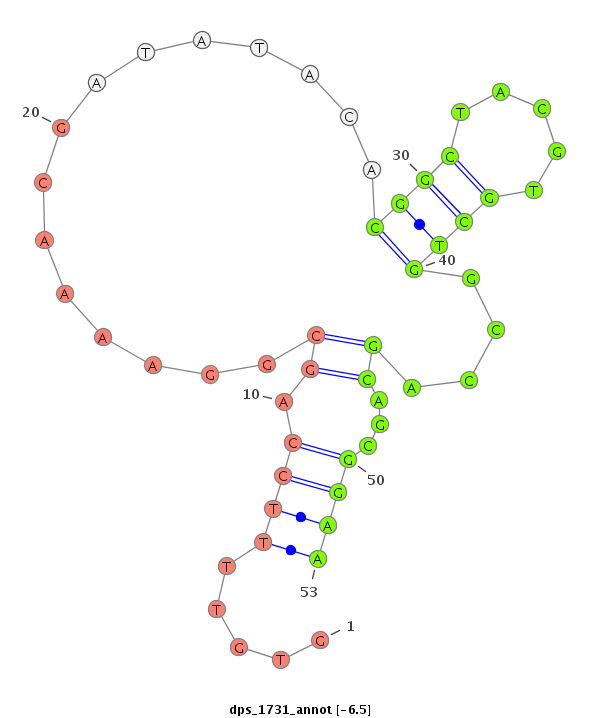
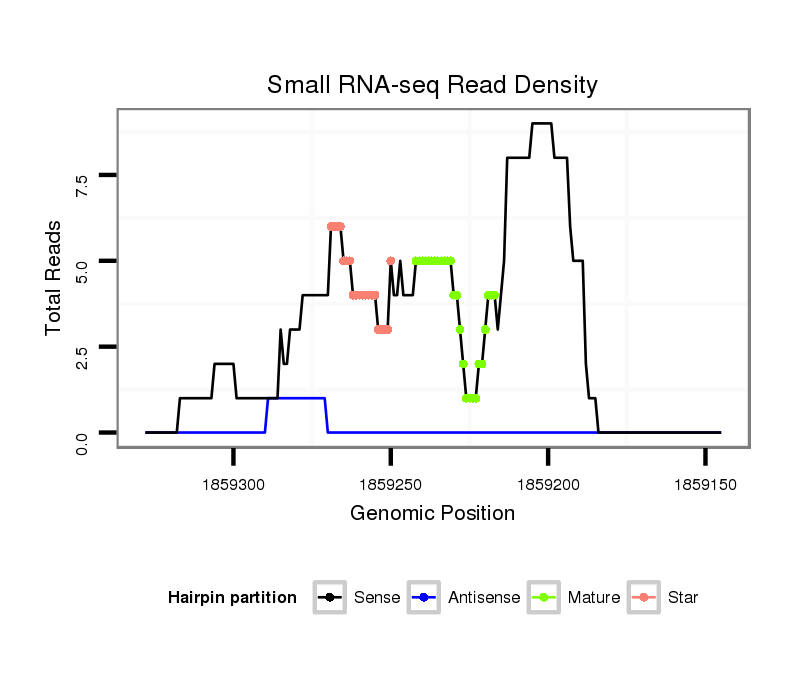
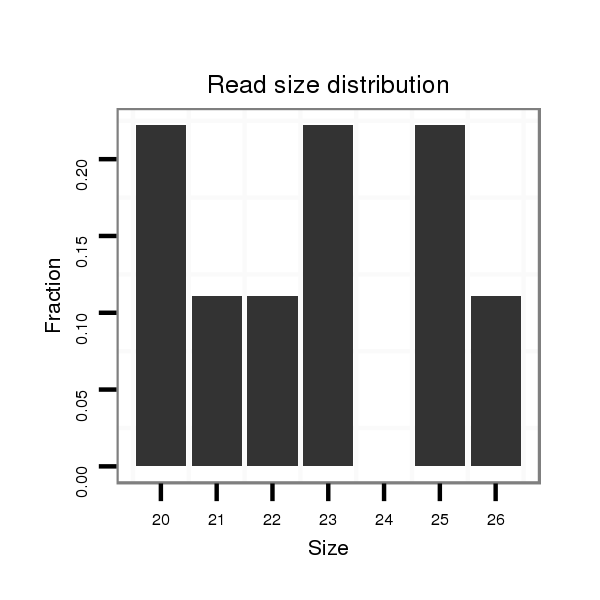
| TCGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGTC-----TA---CGGATT------ACGGCTACGTGCTGGCCACTAAACGAAA-----------------------------------------T------------ACGGC---------------GAGTAAAAGTGGTCGAAGTTCCTCTCCTTC------TG------------CCTCTTCCTCGTCGGCGGAGAGTTCAC------------------------------CCGTTCTGGGTG | Size | Hit Count | Total Norm | Total | M020
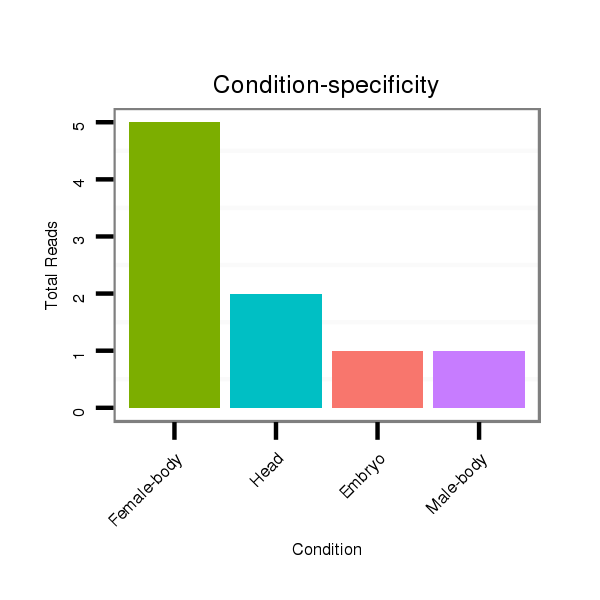
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ..........................................................................................................C-----TA---CGGATT------ACGGCTACGTGCTGGC............................................................................................................................................................................................................. | 25 | 1 | 8.00 | 8 | 0 | 1 | 5 | 1 | 1 |
| ..........................................................................................................C-----TA---CGGATT------ACGGCTACGTGCT................................................................................................................................................................................................................ | 22 | 1 | 6.00 | 6 | 0 | 0 | 3 | 3 | 0 |
| .........................................................................................................................................................GAAA-----------------------------------------T------------ACGGC---------------GAGTAAAAGT............................................................................................................. | 20 | 1 | 3.00 | 3 | 0 | 0 | 3 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................CTCTTCCTCGTCGGCGGAGAGTTCAC------------------------------C........... | 27 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .......................................................................................................CGTC-----TA---CGGATT------ACGGCTACGTGCT................................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 |
| TCGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGTC-----TA---CG....................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................TCTTCCTCGTCGGCGGAGA................................................ | 19 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................CACTAAACGAAA-----------------------------------------T------------ACGGC---------------GAGTA.................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................TC-----TA---CGGATT------ACGGCTACGTGCTGGC............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................AAAAGTGGTCGAAGTTCCTCTCCTT.......................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................GATT------ACGGCTACGTGCTGGC............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................GTCGGCGGAGAGTTCAC------------------------------C........... | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| TCGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGT.................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................AAAGTGGTCGAAGTTCCTCTCCT........................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................C------TG------------CCTCTTCCTCGTCGGC..................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .CGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGTC-----TA---CGGAT.................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................CACTAAACGAAA-----------------------------------------T------------ACGGC---------------G...................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................GGCGGAGAGTTCAC------------------------------CCGTTCTGGG.. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................GAAA-----------------------------------------T------------ACGGC---------------GAGTAAAAGTGGTC......................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GAAGTTCCTCTCCTTC------TG------------CCT.................................................................. | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............CA----------------------TCG---------------------------------------------------------TCTCCATCGTC-----TA---CGGATT------............................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................CACTAAACGAAA-----------------------------------------T------------ACGGC---------------GAGTAAAAGT............................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................GTC-----TA---CGGATT------ACGGCTACGT................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................AACGAAA-----------------------------------------T------------ACGGC---------------GAGTAAAA............................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................ATCGTC-----TA---CGGATT------ACGGC........................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................AAACGAAA-----------------------------------------T------------ACGGC---------------GAGT................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .CGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGT.................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................CGTCGGCGGAGAGTTCAC------------------------------CC.......... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TC------TG------------CCTCTTCCTCGTCGGCGGAGA................................................ | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................AA-----------------------------------------T------------ACGGC---------------GAGTAAAAGTGGTC......................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................CTTC------TG------------CCTCTTCCTCGTCGGCGGA.................................................. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .CGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGTC-----.............................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................GGCCACTAAACGAAA-----------------------------------------T------------ACGGC---------------GA..................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................AAACGAAA-----------------------------------------T------------ACGGC---------------GAGTAAA................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................CATCGTC-----TA---CGGATT------ACGGC........................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| TCGC--------CA----------------------TCG---------------------------------------------------------TCTCCATCGTC-----TA---CGGATT------............................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................AAACGAAA-----------------------------------------T------------ACGGC---------------GAGTAAAAGT............................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................GTC-----TA---CGGATT------ACGGCTACGTGCTGGCC............................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................CTCCATCGTC-----TA---CGGAAC------G............................................................................................................................................................................................................................ | 19 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |