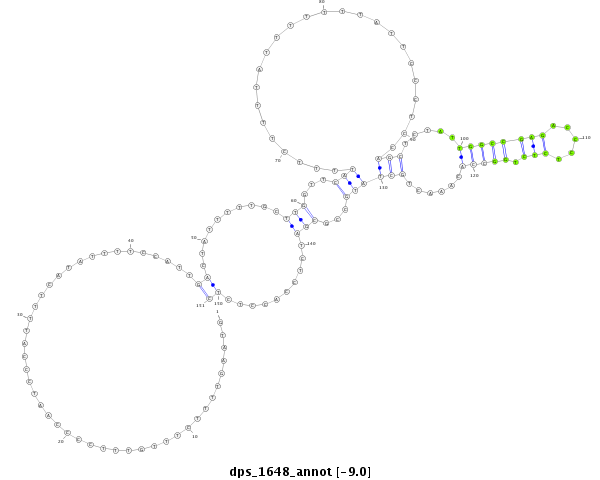
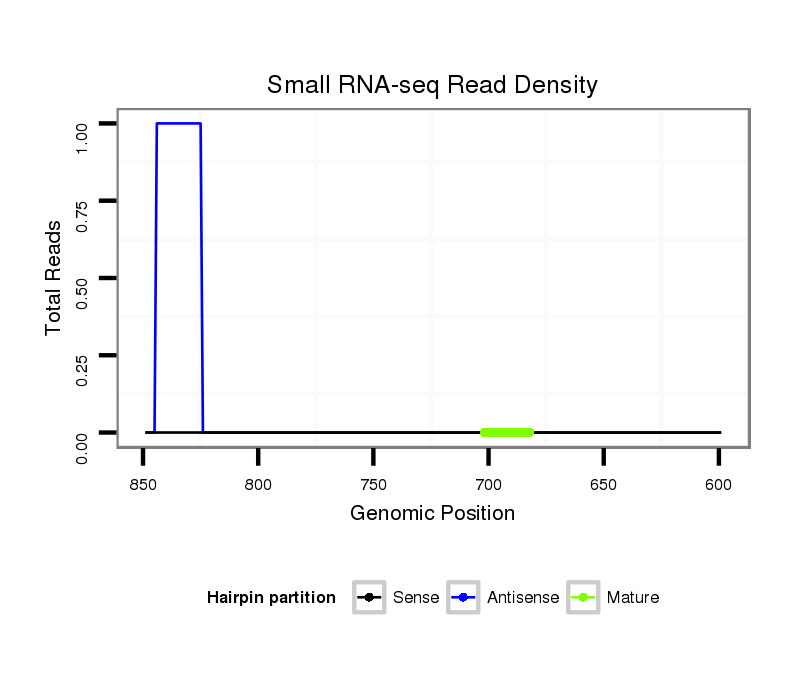
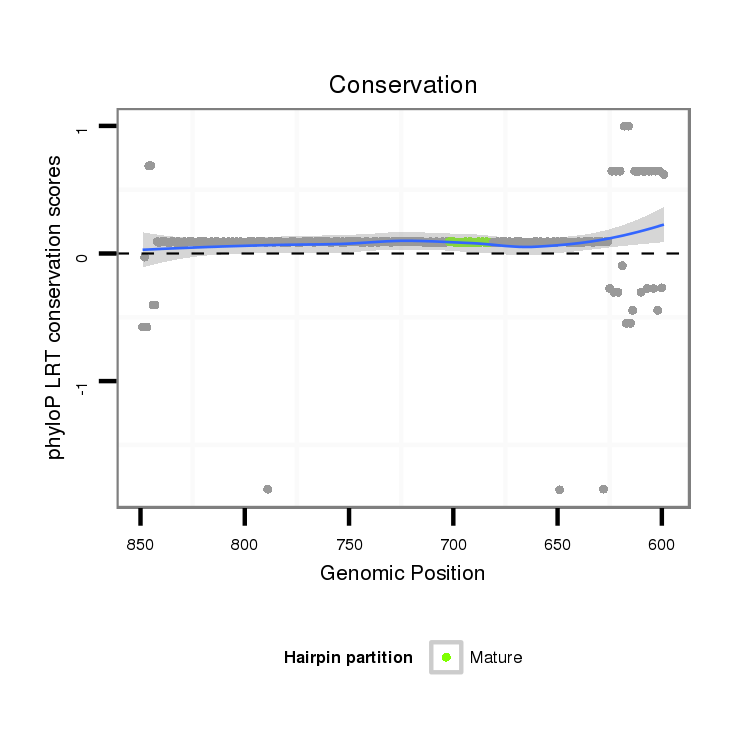
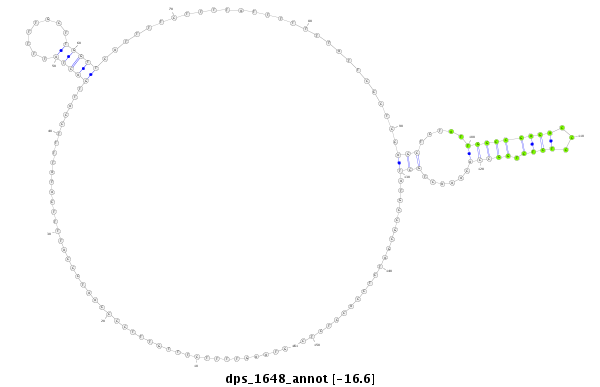
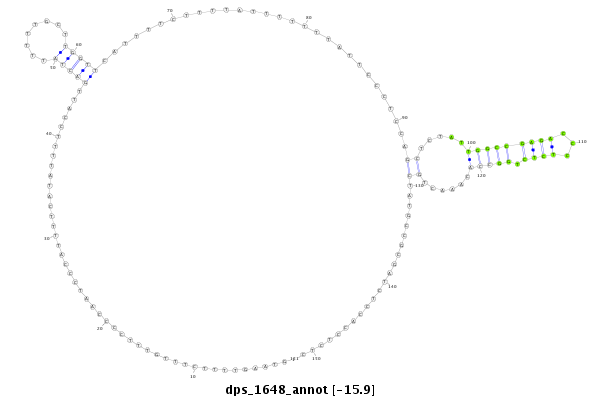
ID:dps_1648 |
Coordinate:4_group4:649-799 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
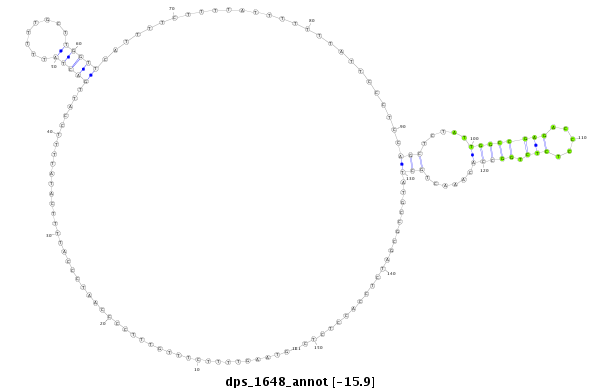
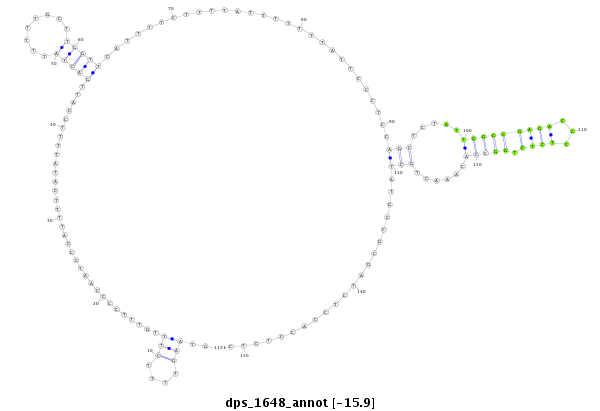
| -16.6 | -15.9 | -15.9 | -15.9 |
 |
 |
 |
 |
CDS [Dpse\GA28927-cds]; exon [dpse_GLEANR_9231:1]; intron [Dpse\GA28927-in]
No Repeatable elements found
| mature | star |
| -----------------------###########################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCGATCGTCTTTCCTTCAGCACAATGCTTTACCTCATCGACTCTGACAGGGTAAGTTTTCTTTGTTTCCCCAATCCCATTTTCATATTTTCCATTGACTATTTTTGCTTGGTTCATTTTCTTTTATTTTTTTATTCCCTCCAGCTCTATTGGCCGAGACCCTCTCTGGCCACAAACTGCTATGCCGCGATCTCCACCTCTCATTCCGCCAGGTAAAGTTATTAAGTTTTTATTTATTATTTACATCATTAC **************************************************.............................................((..........(((...(((.........................(((.....((((((((.....))).)))))......))))))...)))..........))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM444067 head |
GSM343916 embryo |
M040 female body |
|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................TCATACTGCCAGGTAAAGTTGTT............................. | 23 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .........................................................................................................GCTGGGCTAATTTTCTTTTATTT........................................................................................................................... | 23 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ...................................................................................................................................................ATTGGCCGAGACCCTCTCTGG................................................................................... | 21 | 0 | 8 | 0.13 | 1 | 0 | 0 | 1 |
|
CGCTAGCAGAAAGGAAGTCGTGTTACGAAATGGAGTAGCTGAGACTGTCCCATTCAAAAGAAACAAAGGGGTTAGGGTAAAAGTATAAAAGGTAACTGATAAAAACGAACCAAGTAAAAGAAAATAAAAAAATAAGGGAGGTCGAGATAACCGGCTCTGGGAGAGACCGGTGTTTGACGATACGGCGCTAGAGGTGGAGAGTAAGGCGGTCCATTTCAATAATTCAAAAATAAATAATAAATGTAGTAATG
**************************************************.............................................((..........(((...(((.........................(((.....((((((((.....))).)))))......))))))...)))..........))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
GSM343916 embryo |
V112 male body |
M062 head |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....GCAGAAAGGAAGTCGTGTTA.................................................................................................................................................................................................................................. | 20 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................GGGTTAGGGTAAAAGTATAAA.................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................CGAAATGGAGTAGCTGAGAC.............................................................................................................................................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....GCAGAAAGGAAGTCGTGTT................................................................................................................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........AGGAAGTCGTTATACGAAA............................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................GTTACGAAATGGAGTAGCTG.................................................................................................................................................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AAAAAAATAAGGGAGTTCGT.......................................................................................................... | 20 | 2 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CGAGATAACCGGCTCTGGGAG........................................................................................ | 21 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AAAAAAATAAGGGAGGTCAT.......................................................................................................... | 20 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ACAAAGAGGTTAGAGTAA........................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................ATAAGGGAGGTCGAGATAACCG.................................................................................................. | 22 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GGGTAACAGTATATAAGGTG............................................................................................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................GGAAATAAAAAAATAAAGGA................................................................................................................ | 20 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................AGGGTTTAGGGTAACAGTAG..................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................CGAAAAAGTATAATAGGTAA............................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group4:599-849 - | dps_1648 | GCGATCGTCTTTCCTTCAGCACAATGCTTTACCTCATCGACTCTGACAGGGTAAGTTTTCTTTGTTTCCCCAATCCCATTTTCATATTTTCCATTGACTATTTTTGCTTGGTTCATTTTCTTTTATTTTTTTATTCCCTCCAGCTCTATTGGCCGAGACCCTCTCTGGCCACAAACTGCTATGCCGCGATCTCCACCTCTCATTCCGCCAGGTAAAGTTATTAAGTTTTTATTTATTATTTACATCATTAC---- |
| droPer2 | scaffold_6019:844-1094 - | GCGATCGTCTTTCCTTCAGCACAATGCTTTACCTCATCGACTCTGACAGGGTAAGTTTTCCTTGTTTCCCCAATCCCATTTTCATATTTTCCATTGACTATTTTTGCTTGGTTCATTTTCTTTTATTTTTTTATTCCCTCCAGCTCTATTGGCCGAGACCCTCTCTGGCCACAAACTGCTATGCCGCGATCTCCACCTCTTATTCCGCCAGGTAAAGTTATCAAGTTTTTATTTATTATTTACATCATTAC---- | |
| droVir3 | scaffold_13047:8253493-8253504 + | TCCATAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTCTT-------------------- | |
| droKik1 | scf7180000302472:1877085-1877117 + | AT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATATGTATGGTATATATATTAGTGCCTAG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/18/2015 at 01:10 AM