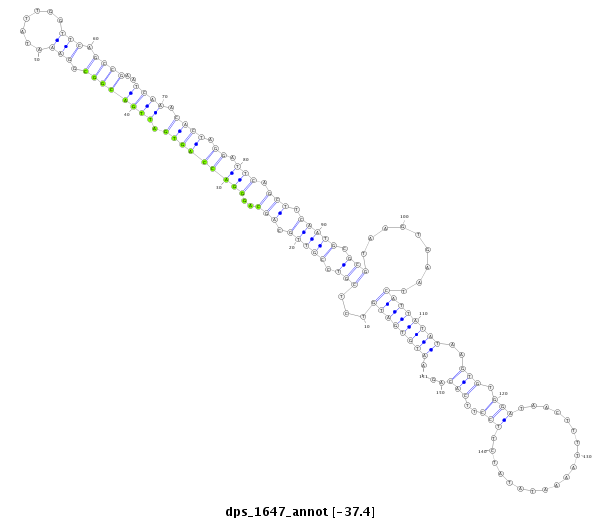
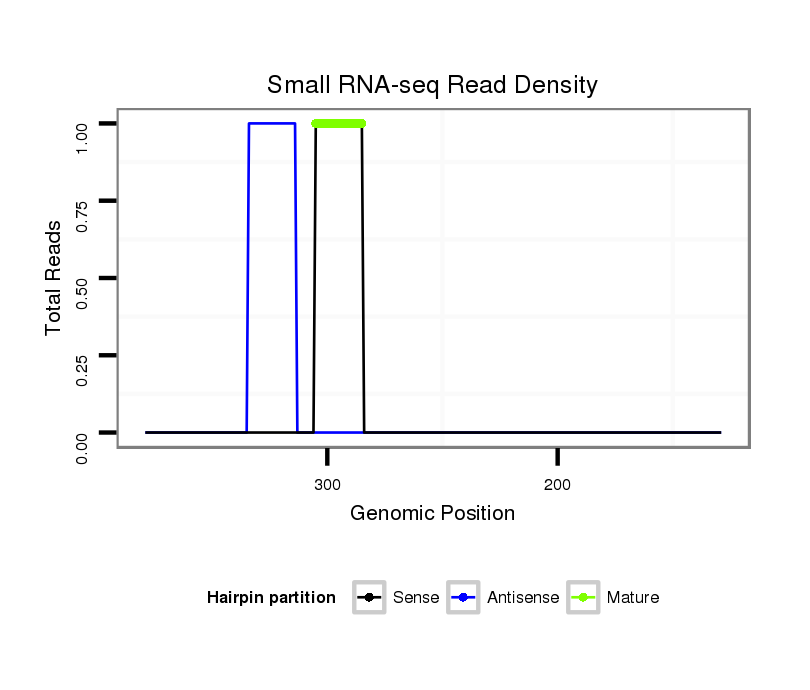
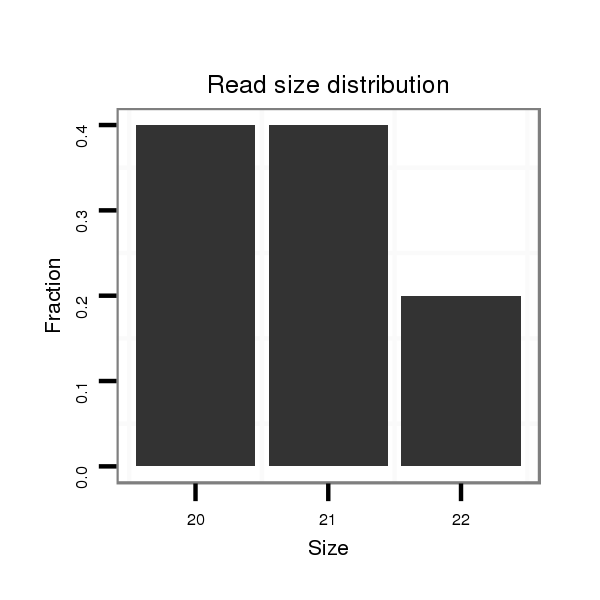
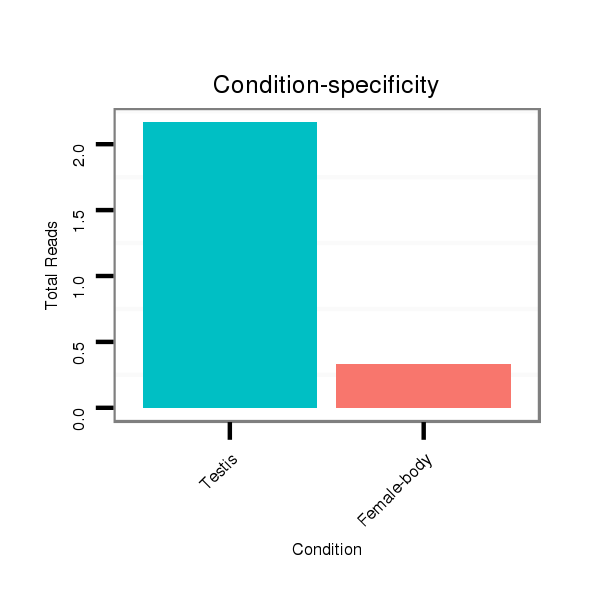
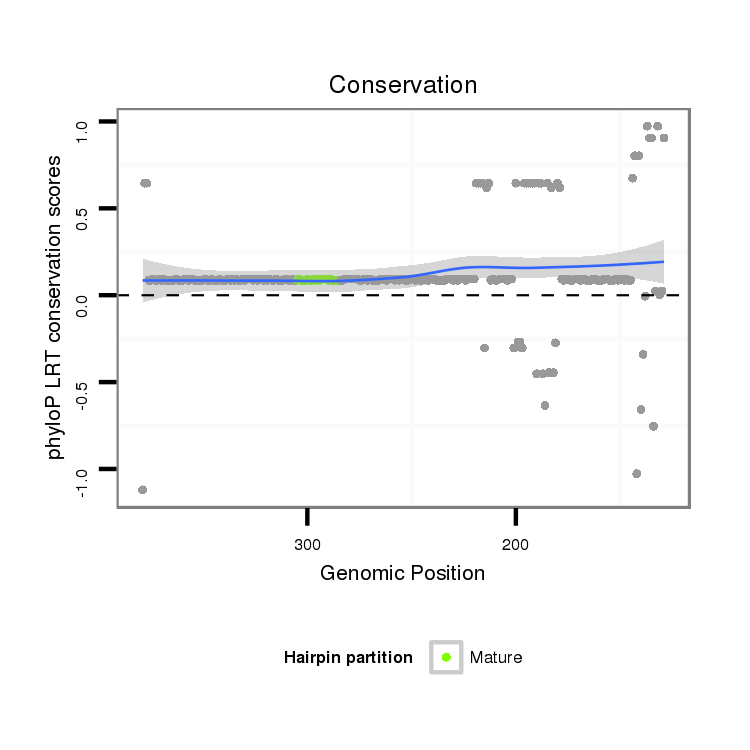
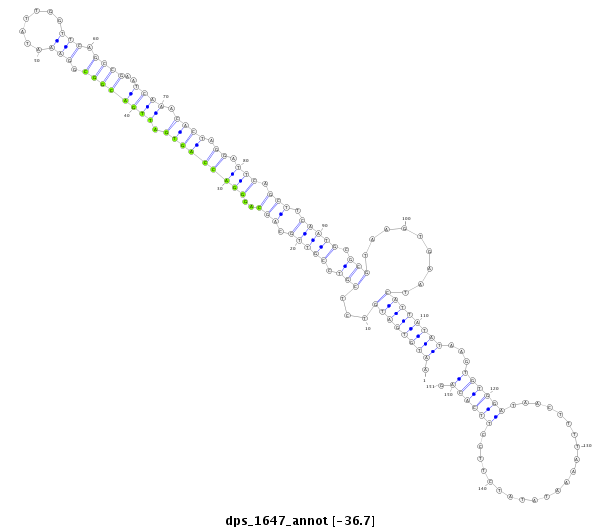
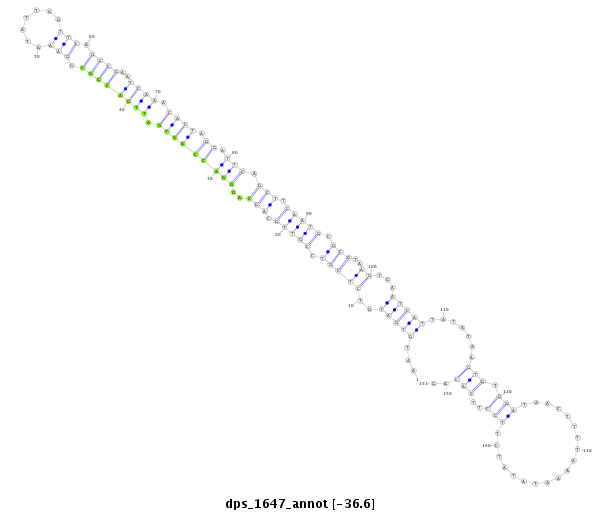
ID:dps_1647 |
Coordinate:4_group4:179-329 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -36.7 | -36.6 | -36.6 |
 |
 |
 |
CDS [Dpse\GA28927-cds]; exon [dpse_GLEANR_9231:2]; intron [Dpse\GA28927-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTAGCTGGACTGGGGCAGGCATCATGCCGCTGCACGGTCGCTGAGACGACAATGTGATGTCTCGTCCGTTGCAGCAGGGACCAGTGATTGACGGCGGAAATATTGGTTCAGCCGAATCAAACACTAGGATTCAGCTTCAATGCGCGTAAGTGAATCATTATATAAGTGTGGATAACTTTTAAAATATATCTTCCTTCACAGAGTACCTATGTACCCTACGCTTTGCCACAAGACGAGTCGTATCCGATGTC **************************************************.((((((((...(((.(((((.(((..(((((((((.((((((((.(((.......))).))))..)))).)))).)).))).))).))))).))).........))))))))..(((.(((...................)))..)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
V112 male body |
M062 head |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TCAGCCAAATCAAACTCTAGA........................................................................................................................... | 21 | 3 | 7 | 15.57 | 109 | 103 | 0 | 6 | 0 | 0 |
| ...........................................................................................................TCAGCCAAATCAAACTCTAG............................................................................................................................ | 20 | 2 | 3 | 15.33 | 46 | 46 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CTCAGCCAAATCAAACTCTAG............................................................................................................................ | 21 | 3 | 9 | 11.11 | 100 | 98 | 0 | 2 | 0 | 0 |
| ............................................................................................................CAGCCAAATCAAACTCTAGAA.......................................................................................................................... | 21 | 3 | 13 | 2.31 | 30 | 30 | 0 | 0 | 0 | 0 |
| .............................................................................................................AGCCAAATCAAACTCTAGAA.......................................................................................................................... | 20 | 3 | 20 | 1.65 | 33 | 33 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCAGCCAAATCAAACTCTAGAA.......................................................................................................................... | 22 | 3 | 4 | 1.50 | 6 | 6 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGGCGGTAACATGGGTTC.............................................................................................................................................. | 19 | 3 | 8 | 1.00 | 8 | 6 | 0 | 2 | 0 | 0 |
| ..........................................................................CAGGGACCAGTGATTGACGGC............................................................................................................................................................ | 21 | 0 | 3 | 1.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| ...........................................................................AGGGACCAGTGATTGACGGC............................................................................................................................................................ | 20 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................TCAGCCAAATCAAACACTAG............................................................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CCACAAGACGAGTCGTATCCGA.... | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................GTACGTTGCAGCAGGGACC......................................................................................................................................................................... | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....TGGACTGGGGCAGGCATC.................................................................................................................................................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................ATCATGCCGCTGCACGGTCGCTGA............................................................................................................................................................................................................... | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGAAGGCGGAAATATTGTTT............................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....CTGGACTGGGGCAGGCATCATGCC............................................................................................................................................................................................................................... | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......GACTGGTTCAGGCATCATTCCG.............................................................................................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GCGGAAATATTGGTTCAGCCCA........................................................................................................................................ | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....CTGGACTGGGGCAGGCATC.................................................................................................................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....TGGACTGGGGCAGGCATCATG................................................................................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................TGATTGACGGCGGAAATATTGG................................................................................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................TAAGATTCAGCATCAATTCGC.......................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................CAGCCAAATCAAACTCTAGG........................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................GGACCAGTGATTGACGGCGG.......................................................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ATCAGCCAAATCAAACTCTAG............................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CACAGGACGGGTCGTGTCCGA.... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................CAGCCAAATCAAACTCTAG............................................................................................................................ | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCAGCCAAATCAAACTCTA............................................................................................................................. | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCAGCCAAATCAAACTCTAGC........................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................CAGCCAAATCAAACTCTAGCA.......................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................GTACCGGTGATTGACGCC............................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
AATCGACCTGACCCCGTCCGTAGTACGGCGACGTGCCAGCGACTCTGCTGTTACACTACAGAGCAGGCAACGTCGTCCCTGGTCACTAACTGCCGCCTTTATAACCAAGTCGGCTTAGTTTGTGATCCTAAGTCGAAGTTACGCGCATTCACTTAGTAATATATTCACACCTATTGAAAATTTTATATAGAAGGAAGTGTCTCATGGATACATGGGATGCGAAACGGTGTTCTGCTCAGCATAGGCTACAG
**************************************************.((((((((...(((.(((((.(((..(((((((((.((((((((.(((.......))).))))..)))).)))).)).))).))).))))).))).........))))))))..(((.(((...................)))..)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M022 male body |
SRR902011 testis |
M059 embryo |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................GTCGGTTTAGTTTGAGATCTT.......................................................................................................................... | 21 | 3 | 13 | 107.46 | 1397 | 1297 | 84 | 16 | 0 | 0 | 0 |
| ...........................................................................................................AGTCGGTTTAGTTTGAGATCT........................................................................................................................... | 21 | 3 | 7 | 102.71 | 719 | 628 | 87 | 4 | 0 | 0 | 0 |
| ...........................................................................................................AGTCGGTTTAGTTTGAGATC............................................................................................................................ | 20 | 2 | 3 | 76.67 | 230 | 190 | 38 | 2 | 0 | 0 | 0 |
| ..........................................................................................................GAGTCGGTTTAGTTTGAGATC............................................................................................................................ | 21 | 3 | 9 | 46.22 | 416 | 354 | 59 | 3 | 0 | 0 | 0 |
| ...........................................................................................................AGTCGGTTTAGTTTGAGA.............................................................................................................................. | 18 | 2 | 20 | 13.50 | 270 | 261 | 2 | 7 | 0 | 0 | 0 |
| ...........................................................................................................AGTCGGTTTAGTTTGAGATCTT.......................................................................................................................... | 22 | 3 | 4 | 11.00 | 44 | 42 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................................TCGGTTTAGTTTGAGATCTT.......................................................................................................................... | 20 | 3 | 20 | 5.70 | 114 | 114 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................AGTCGGTTTAGTTTGAGAT............................................................................................................................. | 19 | 2 | 7 | 5.00 | 35 | 30 | 5 | 0 | 0 | 0 | 0 |
| ............................................................................................................GTCGGTTTAGTTTGAGATC............................................................................................................................ | 19 | 2 | 7 | 4.00 | 28 | 27 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................TCGGTTTAGTTTGAGATC............................................................................................................................ | 18 | 2 | 11 | 1.36 | 15 | 13 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................AAGTCGGTTTAGTTTGAGATC............................................................................................................................ | 21 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................TGCTGTTACACTACAGAGCAG......................................................................................................................................................................................... | 21 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AAGTCGGTTTAGTTTGAGATCT........................................................................................................................... | 22 | 3 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .....................................................CACTACAGAGCAGGCAACGT.................................................................................................................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CCGAGTCGGTTTAGTTTGAGATC............................................................................................................................ | 23 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGGCTTAGTTTGTGATCCTAA........................................................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................CACTACAGAGCAGGCAACGTC................................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................CAGCGACTCTGCTGTTACACT.................................................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GAAACGGTGTTCTGCTCAGC........... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................CGCCTTTATAACCAAGTCGGC......................................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................ACGTGCCAGCGACTCTGCTGT........................................................................................................................................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................ACGGTGTTCTGCTCAGCATAG....... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................CTTTATAACCAAGTCGGCTT....................................................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................GTTACGCGCGTTCACTTAGT.............................................................................................. | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................AAAGTCGGTTTAGTTTGAGATC............................................................................................................................ | 22 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TAGTCGGTTTAGTTTGAGATC............................................................................................................................ | 21 | 3 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGAGTCGGTTTAGTTTGTGAT............................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TCGGTTTAGTTTGAGATCCTT......................................................................................................................... | 21 | 3 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGAGTCGGCTTAGTTTGAGAT............................................................................................................................. | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CGTCGGTTTAGTTTGAGATC............................................................................................................................ | 20 | 3 | 18 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TTTTATATAGAAGGAAGTGT................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................CATGTGATGCGAAACGGTGTC.................... | 21 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TATTGAAATGTTTATATAG............................................................. | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................AGTCGGTTTAGTTTGAGACC............................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................ATACATGTGATGCGAAACGGT....................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group4:129-379 - | dps_1647 | TTAGCTGGACTGGGGCAGGCATCATGCCGCTGCACGGTCGCTGAGACGACAATGTGATGTCTCGTCCGTTGCAGCAGGGACCAGTGATTGACGGCGGAAATATTGGTTCAGCCGAATCAAACACTAGGATTCAGCTTCAATGCGCGTAAGTGAATCATTATATAAGTGTGGATAACTTTTAAAATATATCTTCCTTCACAGAGTACCTATGTACCCTACGCTTTGCCACAAGACGAGTCGTATCCGATGTC |
| droPer2 | scaffold_6019:374-624 - | TTAGCTGGACTGGGGCAGGCATCATGCCGCTGCACGGTCGCTGAGACGACAATGTGATGTCTCGTCCGTTGCAGCAGGGACCAGTGATTGACGGCGGAAATATTGGTTCAGCCGAATCAAACACTAGGATTCAGCTTCAATGCGCGTAAGTGAATCATTATATAAGTGTGGATAACTTTTAAAATATATCTTCCTTCACAGAGTACCTATGTACCCTACGCTTTGCCACAAGACGAGTCGTATCCGATGTC | |
| droKik1 | scf7180000302472:1877085-1877117 + | ATA-------------------------------------------------------------------------------------------------------------------------------------------------------------TATATGT-----------ATGGTATATATATTAGTGCCTAG-------------------------------------------------- | |
| droFic1 | scf7180000449391:947-962 - | A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCGAGTCCTGTGCC | |
| droRho1 | scf7180000777283:2112-2122 - | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCCCGTACC | |
| droTak1 | scf7180000415880:356744-356760 - | G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGCCTATCCCGTGCC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/18/2015 at 01:09 AM