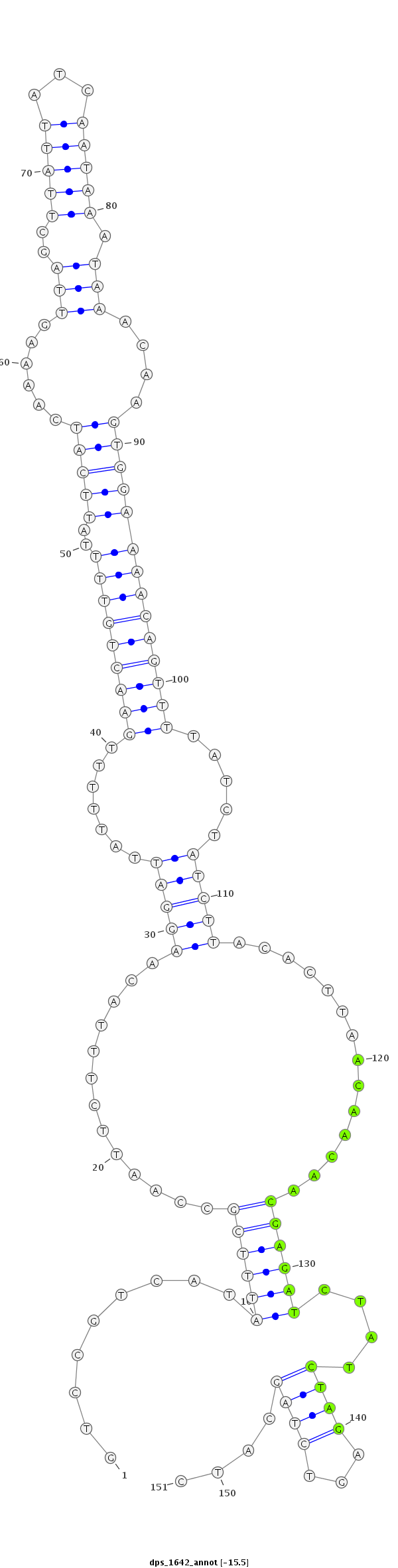
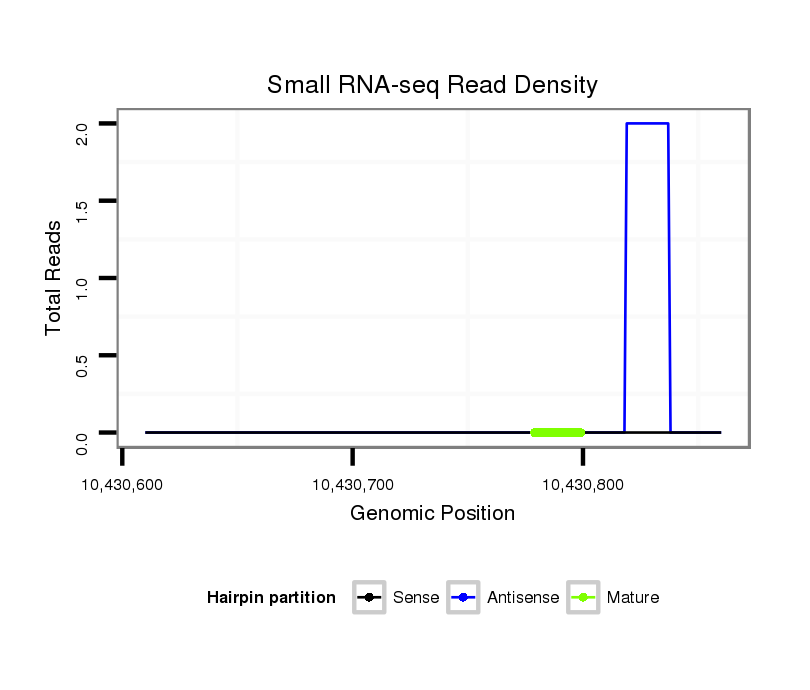
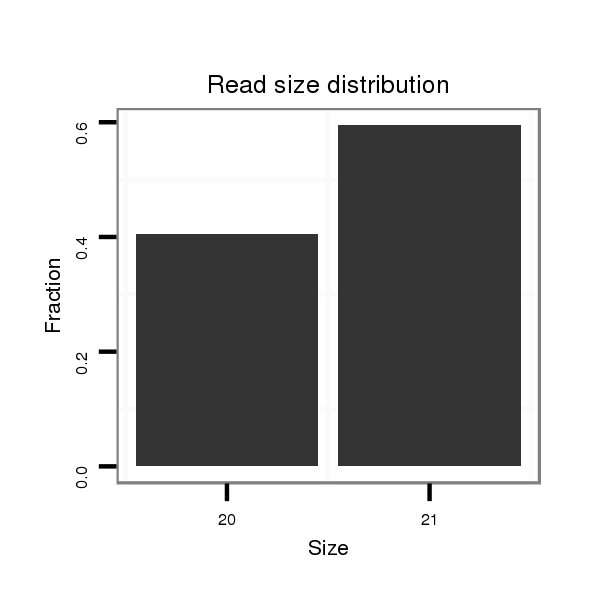
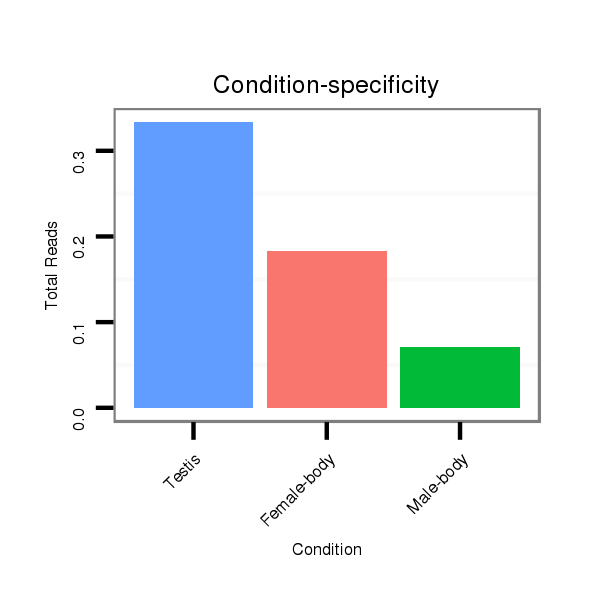
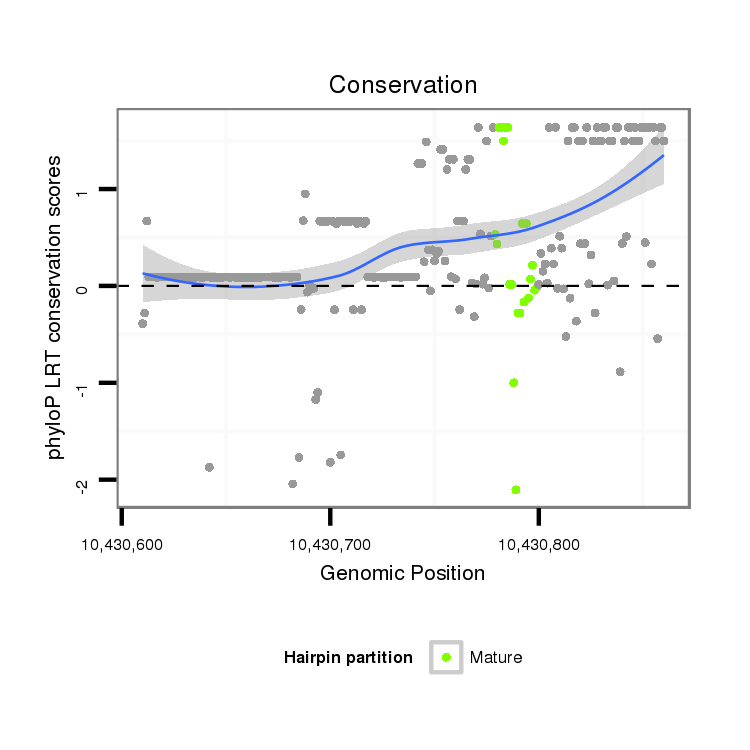
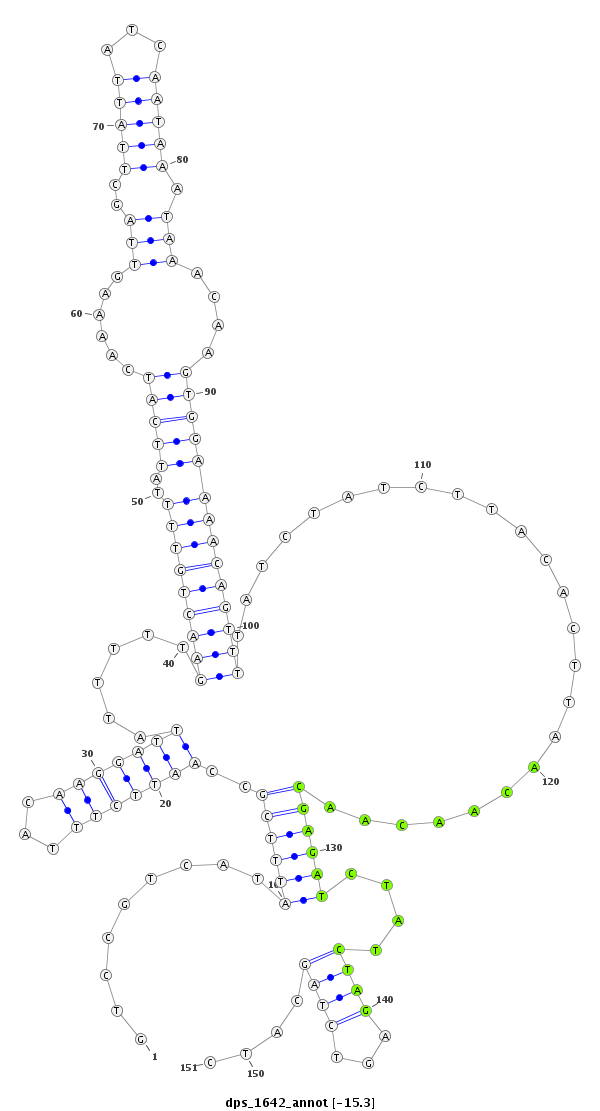
ID:dps_1642 |
Coordinate:4_group3:10430660-10430810 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.3 | -15.3 | -15.1 |
 |
 |
 |
CDS [Dpse\GA26090-cds]; exon [dpse_GLEANR_2798:1]; intron [Dpse\GA26090-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATAATAAGAAAAATATCCCTGATCCCTTGGAATGGACTCAAACAACCCCAGTCCGTCATATTTCGCCAATTCTTTACAAGGATTATTTTTGAACTGTTTTATTCATCAAAAGTTAGCTTATTATCAATAAATAAACAAGTGGAAAACAGTTTTATCTATCTTACACTTAACAACAACGAGATCTATCTAGAGTCTAGCATCCCGCGTAGCTCTTCGCTTGGTTCCCATAGTCAAAGTCCTTGTTGTCGATG **************************************************.........((((((.............(((((.......(((((((((..(((((......(((..(((((...))))).)))....)))))))))))))).....)))))..............))))))....((((...))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
V112 male body |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................CGCGTAGATATGCGCTTGGT............................. | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CATCCCGCGTGGGTCTCCGC.................................. | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................ACAACAACGAGATCTATCTAG............................................................. | 21 | 0 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 |
| ..........................................................................................................................................................................CAACAACGAGATCTATCTAG............................................................. | 20 | 0 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTCCTACAGTCAAATTCCTTG......... | 22 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TCCTACAGTCAAATTCCTTG......... | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................ACAACGAGATCTATCTAGAGT.......................................................... | 21 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................TAGTCAAAGTCCTTGTTGTCG... | 21 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................................................CAACAACGAGATCTATCTAGA............................................................ | 21 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AACGAGATCTATCTAGAGTC......................................................... | 20 | 0 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ..................................GACTCACACAATCCCGGTC...................................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................ATAGTCAAAGTCCTTGTTGTC.... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................CATAGTCAAAGTCCTTGTTG...... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................ATAGTCAAAGTCCTTGTTGTCGAT. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................ATCCCTCGGAATCGAGTCA................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TCAAAGTCCTTGTTGTCGATG | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
TATTATTCTTTTTATAGGGACTAGGGAACCTTACCTGAGTTTGTTGGGGTCAGGCAGTATAAAGCGGTTAAGAAATGTTCCTAATAAAAACTTGACAAAATAAGTAGTTTTCAATCGAATAATAGTTATTTATTTGTTCACCTTTTGTCAAAATAGATAGAATGTGAATTGTTGTTGCTCTAGATAGATCTCAGATCGTAGGGCGCATCGAGAAGCGAACCAAGGGTATCAGTTTCAGGAACAACAGCTAC
**************************************************.........((((((.............(((((.......(((((((((..(((((......(((..(((((...))))).)))....)))))))))))))).....)))))..............))))))....((((...))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902012 CNS imaginal disc |
SRR902011 testis |
M040 female body |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................GAGAAGCGAACCAAGGGTA....................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................................TTGTTGTTGCTCTAGATAGAT.............................................................. | 21 | 0 | 13 | 0.31 | 4 | 0 | 4 | 0 | 0 |
| ........................................................................................................................................................................CTGTTGTTGCTCTAGATAGATC............................................................. | 22 | 1 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 |
| ................................................................................................................................................................................GCTCTAGATAGATCTCAGATC...................................................... | 21 | 0 | 15 | 0.13 | 2 | 0 | 2 | 0 | 0 |
| ........................................................................................................................................................................TTGTTGTTGCTCTAGATAG................................................................ | 19 | 0 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 |
| ................................................................................................................................................................................................................................GGTATCAGTTTCAGGAACAAC...... | 21 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| .....................................................................................................................................................................................................................................CAGTTTCAGGAACAACAGCTA. | 21 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| ..........................................................................................................................................................................GTTGTTGCTCTAGATAGATCT............................................................ | 21 | 0 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................TTGTTGCTCTAGATAGATCTC........................................................... | 21 | 0 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 |
| ...............AGGGACGAGGAAACCTTA.......................................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................GTTGTTGCTCTAGACAGATCT............................................................ | 21 | 1 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................TTGCTCTAGATAGATACCAGAT....................................................... | 22 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................GCTCTAGATAGATCTCAGAT....................................................... | 20 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................ATCAGTTTCAGGAACAACAGC... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...............................................................GCGGTTAAGAACTGTTCTA......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................TATCAGTTTCAGGAACAACAGCTA. | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................TTGTTGTTGCTCTAGATA................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group3:10430610-10430860 + | dps_1642 | ATAATAAGAAAAATATCCCTGATCCCTTGGAATGGACTCAAACAACCCCAGTCCGTCATATTTCGCCAATTCTTTACAAGGATTA--TTTTTGAACTGTTTTATTCATCAAAAGTTAGCTTATTATCAATAAAT------------------------------------------------------------------AAACAAG--------TGGAAAACAGTTTTATCTAT------------CTTACACTTAACAACAA---CGAGATC-----------------------------------TATCTA--GAGT-CTAGCATCCCGCGTAGCTCTTCGCTTGGTTCCCATAGTCAAAGTCCTT------GTTGTCGATG |
| droPer2 | scaffold_8:1644155-1644405 + | ATAATAAGAAAAATATCCCTGATCCCTTGGAAAGGACTCAAACAACCCCAGTCCGTCATATTTCGCCAATTCGTTTCAAGGATTA--TTTTTTAACTATTTTATTCATCAAAAGTTAGCTTATTATCAATAAAT------------------------------------------------------------------AAACAAG--------TGGAAAACAGTTTTATCTAT------------CTTACACCTAACAACAA---CGAGATC-----------------------------------TATATA--GAGT-CTAGCATCCCGCGTAGCTCTTCGCTTGGTTACCATAGTCAAAGTCCTT------GTTGTCGATG | |
| droAna3 | scaffold_13334:572515-572545 + | A-----------------------------------------------------------------------------AGGATCC--TTTTTGAGCTATTTTACTCACCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droBip1 | scf7180000396728:2217046-2217204 + | AAA-----------------------------------------------------------------------------GTCGA-T-----------------------------------------------TCAT-AAATAAT-------------------CAAAAA---------------------------------AGG--------AGGAAAACAGTATCATCTATCCTAGTAAGAAGTCTAAACCTAACAACAA---CAGCTACC----------------------------------T-ACTG--AGGT-CCATCAGTAATCGTAGCCCCTTCCCTGGTTGCCGTAGCCAAAGTCCTTCAGGTAGTTGTCTATG | |
| droKik1 | scf7180000302472:1879828-1879982 - | A-------------------------------------------------------------------------------------------------------------------------------------A--T-AAATAATTAAATAGTCATAG--------------------------------------------------------------------------------------AACCTAAACCTAACAACAA---CGAT---TGAAACGGAAAGCTCCATTTCCATCTCTTCATCCCCATCTC--TCGC-TCAGTAATCTCCGTAACCCCTGCCTTGGTTGCCATAGCCAAAGTCCTTCAGATAGTTATCTATG | |
| droFic1 | scf7180000449391:678-868 + | T-------------------------------------------------------------------------------------------------------------------------------------TTTT-AAATAATTTAATCATCATGAAGTCAACAGGAGTGGAGACCGTT-A----------ACAAATAAATAAGAGGACACCTAGAAAACATTT----CTAGCCTAGAAGTACGCCTAAACCTAACAACAA---AGAG---C-------------------------------------CTG--GAGC-TCAGTAGTCCGCGTATCCCCTTCCCTGGTTGCCGTATCCAAAGTCCTTCAGGTAGTTATCGATG | |
| droEle1 | scf7180000491186:2533209-2533378 - | T-------------------------------------------------------------------------------------------------------------------------------------TTTT-AAATATTTTAATAATCATTAAGTCAACAG---------------CTTTACTTTATATAAATAAATAAAAGGATCCCTAGGAA----------------------TAAGTCTTAACCTAACAACAACAACGAG---T---------------------------------------G--GAAC-TCAGTAGTCCACATAGCCCCTTCCCTGGTTGCCATAACCAAAGTCCTTCAGGTAGTTATCGATG | |
| droBia1 | scf7180000302408:3629032-3629171 + | A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAATAAATAAGAGGACACCTAGAAAACATTT----CTAGCCTAGAGATACGCCTAAACCTAGCAACAA---TGCA---T---------------------------------------G--GAAC-TTAGTAGTCCGCGTAGCCCCTTCCCTGGTTGCCATAGCCAAAGTCCTTCAGGTAGTTATCAATG | |
| droTak1 | scf7180000415880:356482-356665 + | T--------------------------------------------------------------------------ATAAAAATCTTT-----------------------------------------------TTTT-AAATAGTTTAATCATCATTAGTTCAACAGGGAAGGAGAAAGAA-C----------ATAAATAAATAAGAGGACACCTAT----------------------CGATAAGCCTAAACCTAACAACAA---CGAT---T--------------------------------------CG--GAAC-TCAGTAGTCCGCATAGCCCCTTCCCTGGTTGCCATAACCAAAGTCCTTCAGATAGTTATCAATG | |
| droSec2 | scaffold_3:4994556-4994690 - | A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAATAAATAATGGGATACCTAGAAAGCATT-----CTAGCCTAG-------ACTAATCCTAAAAACAA---CGTG---T---------------------------------------GGGGAACTTCAGTAGTCCGCATATCCCCTTCCTTGGTTGCCATATCCGAAGTCCTTCAGATAATTATCGATG | |
| droYak3 | 2L:12222990-12223171 - | T-------------------------------------------------------------------------------------------------------------------------------------TTTTAAAATAATTTAATCATCATTTAGTCAACAGGAAAGGGGGA-G-TAC-----------TCAATAAATAATGGGATACCTAGAAAACATT-----CTAGCCTAGA------CTTATGCTAAAAAACAA---CAGA---T---------------------------------------G--GAACTTCAGTAGTCCGCATAGCCCCTTCCTTGGTTGCCATATCCAAAGTCCTTCAGGTAATTATCGATG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/18/2015 at 01:09 AM