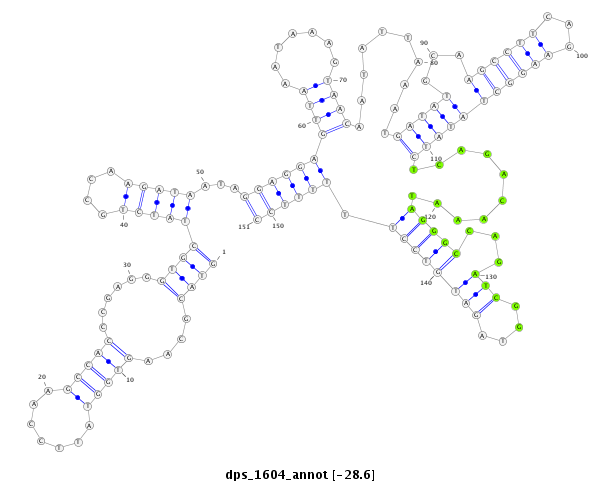



ID:dps_1604 |
Coordinate:4_group3:7423135-7423285 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
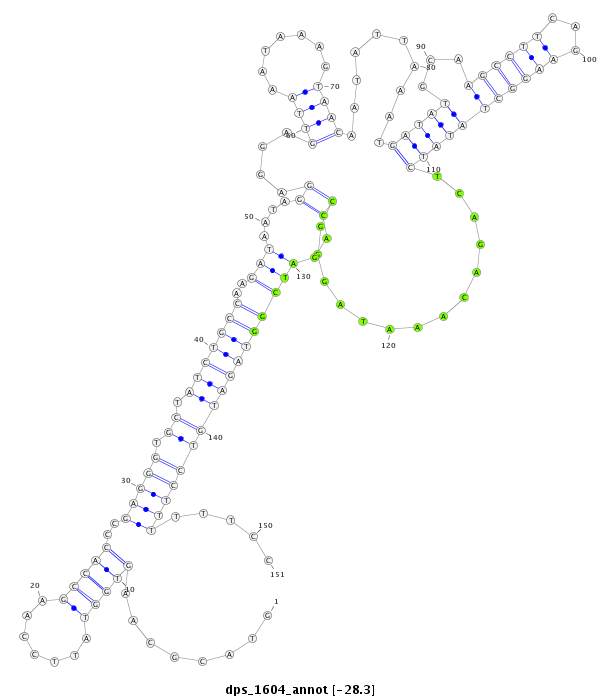
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
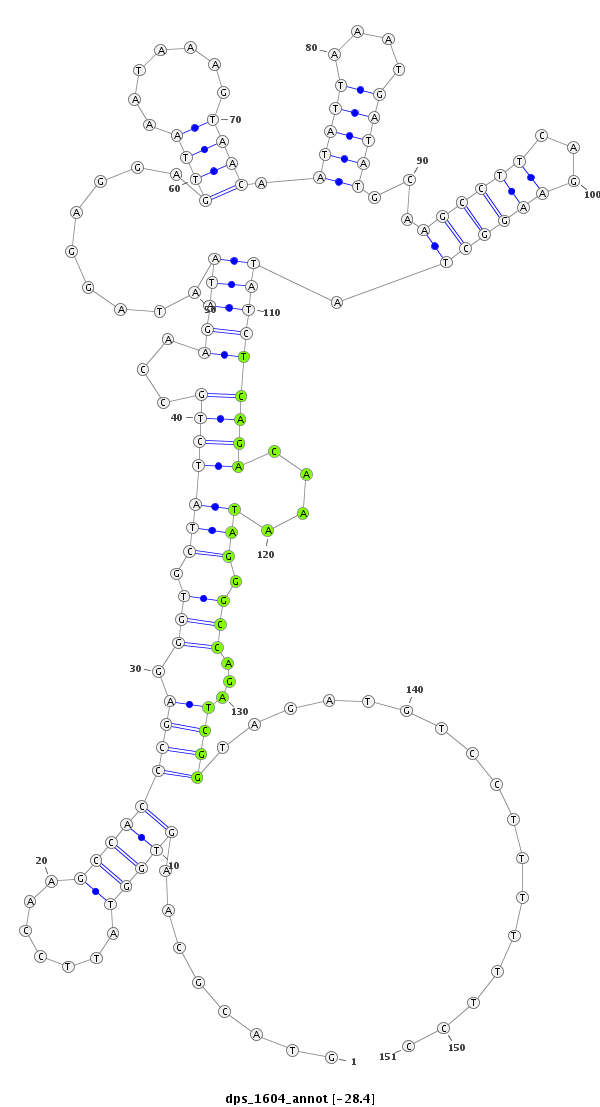
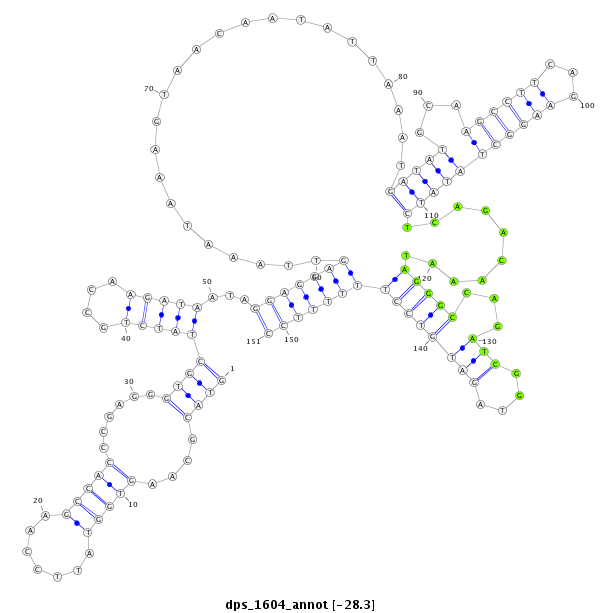
| -28.4 | -28.3 | -28.3 |
 |
 |
 |
CDS [Dpse\GA12955-cds]; exon [dpse_GLEANR_2628:1]; intron [Dpse\GA12955-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAAAAGGGTGATGCCCCTGGTCAACGATGGAGGGCATATCGGTCTCCGGGGTACGCAAGTGGTATTCCAAGCCACCCGAGGGTGCTATCTGCCAAGATAATAGGAGGAGTTAAATAAAGTAACAATATTAAATGATATGCAAGCCTTCAGAAGGCTATATCTCAGACAAATAGGGCCAGATCGGTAGATGTCCTTTTTTCCGATGGCAGTCGACTGGCAGGACTCTCCCGATCACGAAAAGACATGCCGCA **************************************************((((....(((((.......)))))......))))(((((....)))))...((((((((((.......))))..........(((((...((((((...)))))))))))..........(((((...(((....)))))))).))))))************************************************** |
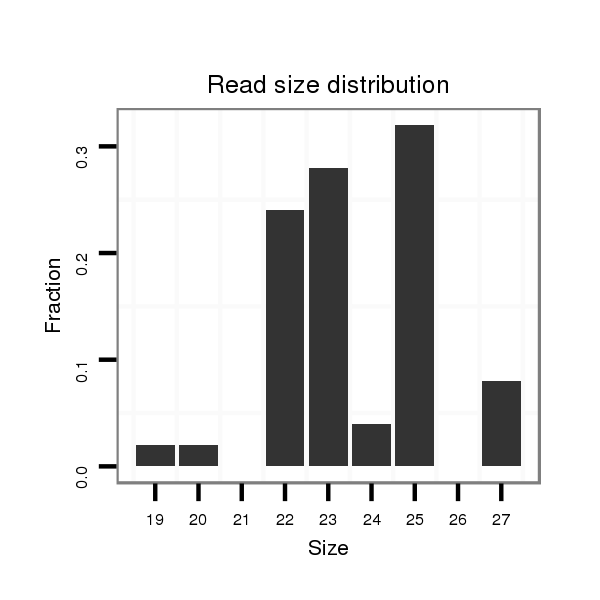
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
GSM343916 embryo |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TCTCAGACAAATAGGGCCAGATCGAGAG................................................................ | 28 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................TCAGACAAATAGGGCCAGATCGG................................................................... | 23 | 0 | 20 | 0.65 | 13 | 0 | 13 | 0 | 0 |
| ...........................................................................................................................................................TATATCTCAGACAAATAGGGCCAGA....................................................................... | 25 | 0 | 20 | 0.65 | 13 | 13 | 0 | 0 | 0 |
| ...............................................................................................................................................................TCTCAGACAAATAGGGCCAGAT...................................................................... | 22 | 0 | 20 | 0.60 | 12 | 12 | 0 | 0 | 0 |
| ..........................ATGGTTAGCATATCGGTCT.............................................................................................................................................................................................................. | 19 | 3 | 15 | 0.47 | 7 | 7 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TCTCCCGATCACGAAAAGACATGCCG.. | 26 | 0 | 20 | 0.25 | 5 | 0 | 3 | 2 | 0 |
| .............................................................................................................................................................TATCTCAGACAAATAGGGCCAGATCGG................................................................... | 27 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 |
| ......................................................................................................................................................................................................TCCGATGGCAGTCGACTGGCA................................ | 21 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ................................................................................................................................................................CTCAGACAAATAGGGCCAGATCGGT.................................................................. | 25 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ..........................ATGGTTAGCATATCGGTCTC............................................................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................TCAGACAAATAGGGCCAGATCGGT.................................................................. | 24 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| .............................................................................................................................................................................................................................ACTCTCCCGATCACGAAAAGACAT...... | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................CAAACCACGCGAGGGTTC...................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TCGACTGGCAGGACTCTCCCGA.................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TCCTTTTTTCCGATGGCAGT......................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................CTCCCGATCACGAAAAGACATGCCGC. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................ATATCTCAGACAAATAGGGCCAG........................................................................ | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................TCTCAGACAAATAGGGCCAGATT..................................................................... | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................TCTCAGACAGATAGGGCCAGATCGG................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................CCTTTTTTCCGATGGCAGTCGA...................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................TCTCAGACAAATAGGGCCAG........................................................................ | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................TATCTCAGACAAATAGGGC........................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............CATGTTGAACGATGGAGGG......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................TGGCAGGACTCTCCCGATCACGAAAA........... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................CCGATCACGAAAAGACATGCC... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TCCTTTTTTCCGATGGCAGTC........................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTCTCAGACAAATAGGGCCAGATC..................................................................... | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................GACTGGCAGGACTCTCCCGA.................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................CCTCAGACAAATAGGGCCAGATCGG................................................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................CTGGCAGGACTCTCCCGATCACGA.............. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
GTTTTCCCACTACGGGGACCAGTTGCTACCTCCCGTATAGCCAGAGGCCCCATGCGTTCACCATAAGGTTCGGTGGGCTCCCACGATAGACGGTTCTATTATCCTCCTCAATTTATTTCATTGTTATAATTTACTATACGTTCGGAAGTCTTCCGATATAGAGTCTGTTTATCCCGGTCTAGCCATCTACAGGAAAAAAGGCTACCGTCAGCTGACCGTCCTGAGAGGGCTAGTGCTTTTCTGTACGGCGT
**************************************************((((....(((((.......)))))......))))(((((....)))))...((((((((((.......))))..........(((((...((((((...)))))))))))..........(((((...(((....)))))))).))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
SRR902010 ovaries |
GSM343916 embryo |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................TCGTGGTCTAGCCATCGACA............................................................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..TTTCCCACTTCGCGGACCAGT.................................................................................................................................................................................................................................... | 21 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGACCGTCCTGAGAGGGCTAGTGCTT............. | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| .......................................................................................AGGCGGTTCTATTACCCTA................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................................TCCTGAGAGGGCTAGTGCTT............. | 20 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AGGCTACCATCAGATGACCGTCCTG............................ | 25 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AGTCTTCCGATATAGAGTCTGTTT................................................................................. | 24 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GACCGTCTTGAGAGGGCTAGTGCTT............. | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................CTCATCAATTGATTTCATA................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................CCTGAGAGGGCTAGTGCTT............. | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CGACCTGAGAGGGCTAGTGCTT............. | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCCCGTCAGCTGACCGTCCTG............................ | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group3:7423085-7423335 + | dps_1604 | CAAAAGGGTGATGCCCCTGGTCAACGATGGAGGGCATATCGGTCTCCGGGGTACGCAAGTGGTATTCCAAGCCACCCGAGGGTGCTATCTGCCAAGATAATAGGAGGAGTTAAATAAAGTAACAATATTAAATGATATGCAAGCCTTCAGAAGGCTATATCTCAGACAAATAGGGCCAGATCGGTAGATGTCCTTTTTTCCGATGGCAGTCGACTGGCAGGACTCTCCCGATCACGAAAAGACATGCCGCA |
| droPer2 | scaffold_1440:4551-4801 + | CAAAAGGGTGATGCCCCTGGTCAACGATGGAGGGCATGTCGGTCTCCGGGGCACGCAAGTGGTATTTCAAGCCACCCAAGGGTGCGATCTGCCAAGATAATAGGAAGAGTTAAATAAAGTAACAATATTAAATGATATGCAAGCCTTCAGAAGGCTATATCTCAGACAAATAGGGCCAGATCGGTAGATGTCCTTTTTTCCGATGGCAGTCGACTGGCAGGACTCTCCCGATAACGAAAAGACATGCCGCA | |
| droAna3 | scaffold_13334:335382-335405 - | C-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGATTCCGAAAATGTACGCCGTT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
Generated: 05/18/2015 at 01:04 AM