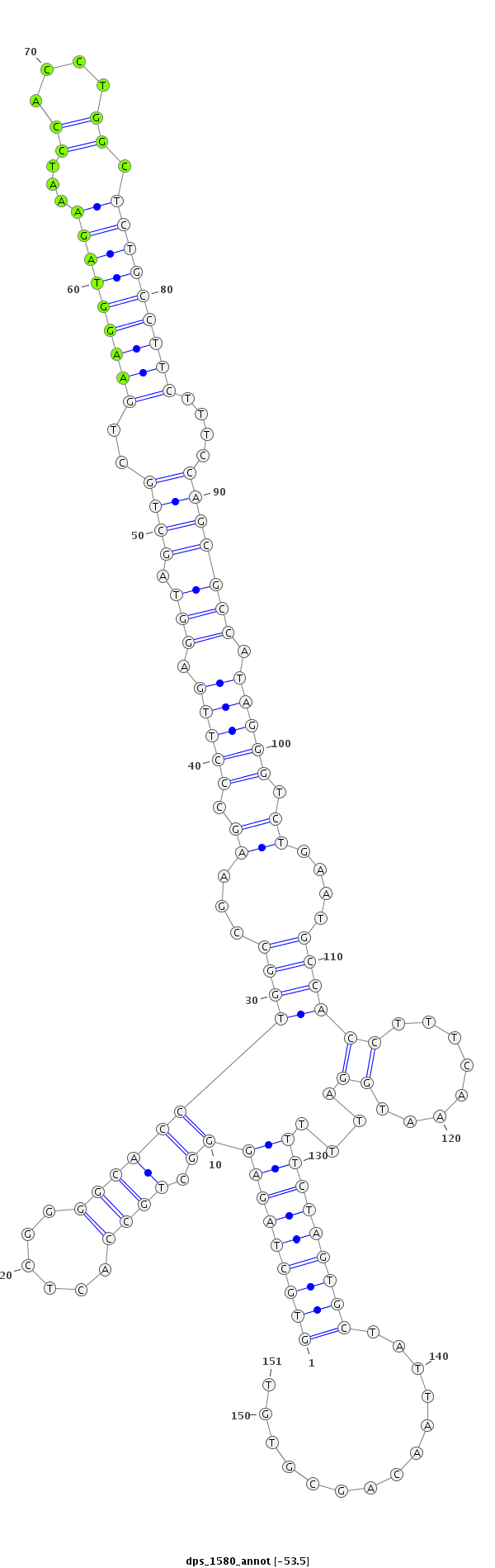
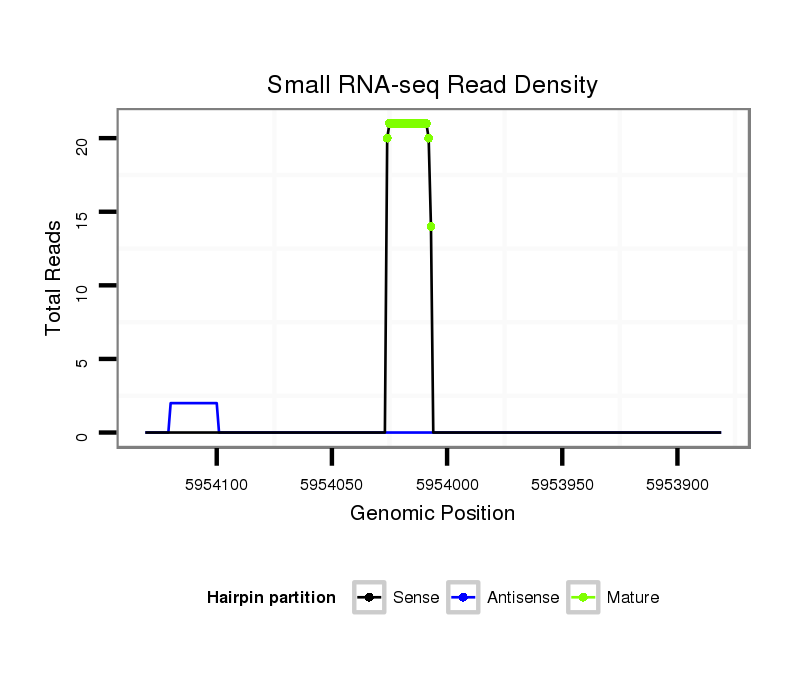
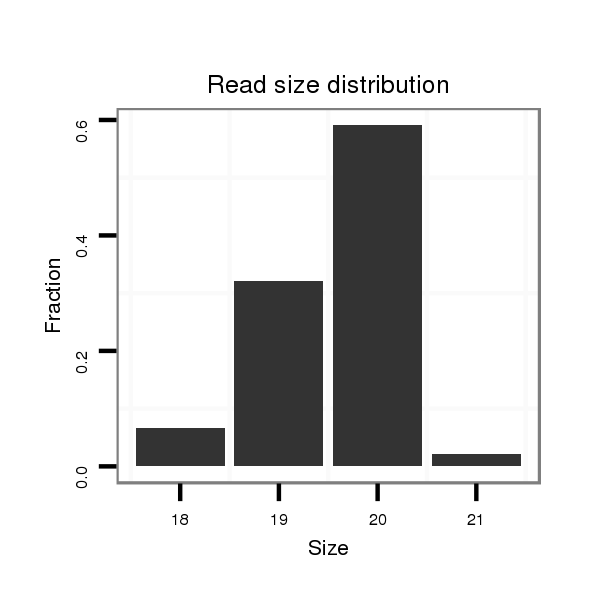
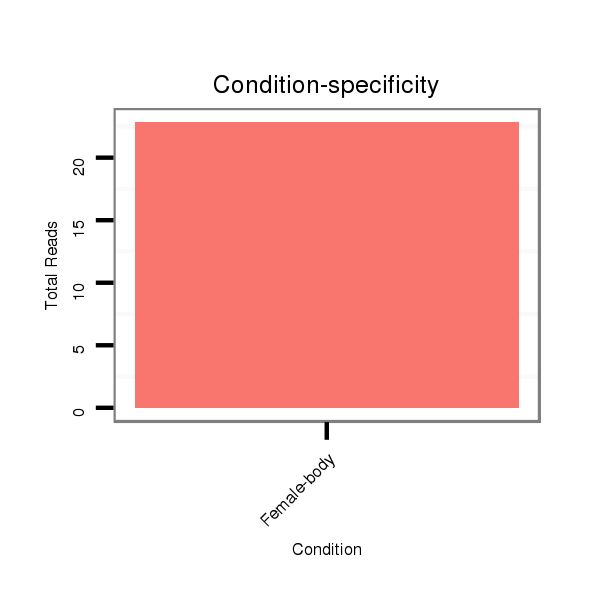
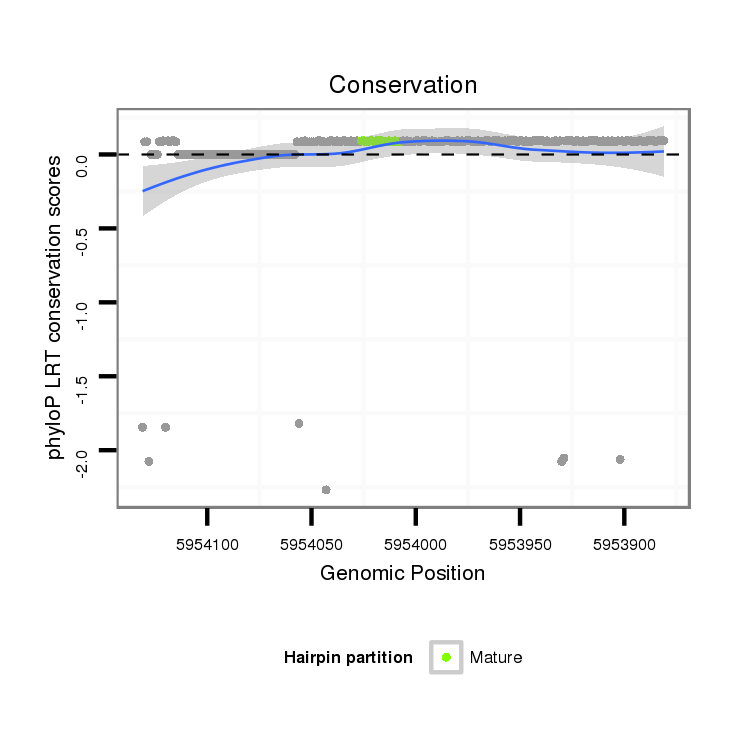
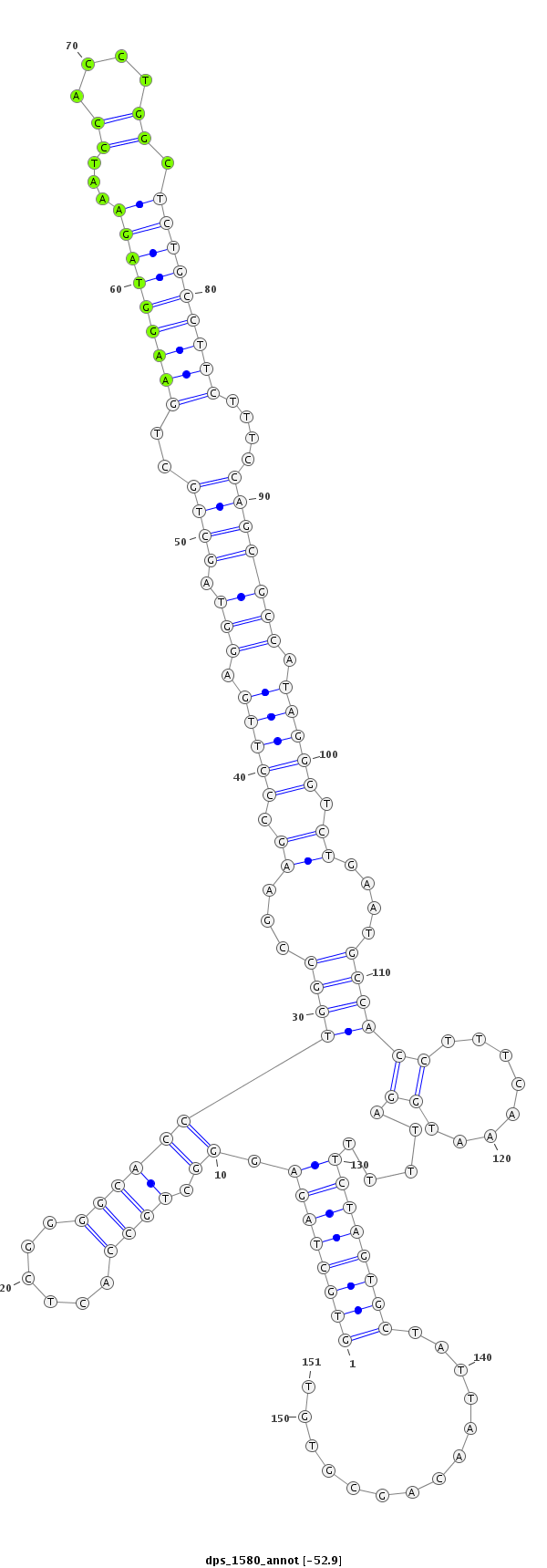
ID:dps_1580 |
Coordinate:4_group3:5953931-5954081 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -52.9 | -52.9 | -52.7 |
 |
 |
 |
exon [dpse_GLEANR_1901:1]; CDS [Dpse\GA25724-cds]; intron [Dpse\GA25724-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TCCACTGGGGTTTCTTCGCTGATTCTGGCCTACAGCATCCGACGCTTATGGTGCTAGAGGGCTGCCACTCGGGGCACCTGGCCGAAGCCCTTGAGGTAGCTGCTGAAGGTAGAAATCCACCTGGCTCTGCCTTCTTTCCAGCGCCATAGGGTCTGAATGCCACCTTTCAAATGGATTTTTCTAGTGCTATTAACAGCGTGTACTTTTCTACTCGTATTTCCTCAGACCATAGAATGCCATGGAATACATTT **************************************************(((((((((((.((((......))))))((((...((.(((((.(((.((((..(((((((((...((....)).)))))))))....))))))).))))).))....))))((........))....)))))))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
SRR902011 testis |
V112 male body |
GSM444067 head |
M062 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................AAGGTAGAAATCCACCTGGC.............................................................................................................................. | 20 | 0 | 6 | 13.50 | 81 | 81 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTGG............................................................................................................................... | 19 | 0 | 6 | 6.33 | 38 | 38 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................TAAGGTAGAAATCCACCTGGCAC............................................................................................................................ | 23 | 2 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................TGTCGAAGGTAGAAATCCACC.................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTG................................................................................................................................ | 18 | 0 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AGGTAGAAATCCACCTGGC.............................................................................................................................. | 19 | 0 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTGGCCC............................................................................................................................ | 22 | 1 | 5 | 0.80 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTGGCAA............................................................................................................................ | 22 | 2 | 6 | 0.67 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTGGCA............................................................................................................................. | 21 | 1 | 6 | 0.67 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GAAGGTAGAAATCCACCTGGCC............................................................................................................................. | 22 | 1 | 5 | 0.60 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTGGCT............................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GAAAGCTACCTGGCTCTGAC........................................................................................................................ | 20 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CAAGGTAGAAATCCACCTGGC.............................................................................................................................. | 21 | 1 | 6 | 0.33 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AGGTAGAAATCCACCTGG............................................................................................................................... | 18 | 0 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CTTTCCGGCGACATATGGTC.................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................CGAAGGTAGAAATCCACCTGGCC............................................................................................................................. | 23 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCGCCTGGC.............................................................................................................................. | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TTAAGGTAGAAATCCACCTGG............................................................................................................................... | 21 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AGGTAGAAATCCACCTGGCA............................................................................................................................. | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GGTAGAAATCCACCTGGC.............................................................................................................................. | 18 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................GGAAGGTAGAAATCCACCTGG............................................................................................................................... | 21 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTAGAAATCCACCTGGA.............................................................................................................................. | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AAGGTACAAATCCACCTGGC.............................................................................................................................. | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........GTTTCATCGTTGATCCTGGC.............................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................GATGTAGCTGATGAAGATAGA.......................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................GCACTTGGCGGAAGGCCTTG.............................................................................................................................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AGGTGACCCCAAAGAAGCGACTAAGACCGGATGTCGTAGGCTGCGAATACCACGATCTCCCGACGGTGAGCCCCGTGGACCGGCTTCGGGAACTCCATCGACGACTTCCATCTTTAGGTGGACCGAGACGGAAGAAAGGTCGCGGTATCCCAGACTTACGGTGGAAAGTTTACCTAAAAAGATCACGATAATTGTCGCACATGAAAAGATGAGCATAAAGGAGTCTGGTATCTTACGGTACCTTATGTAAA
**************************************************(((((((((((.((((......))))))((((...((.(((((.(((.((((..(((((((((...((....)).)))))))))....))))))).))))).))....))))((........))....)))))))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
GSM444067 head |
V112 male body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........AAGAAGCGACTAAGACCGGAT........................................................................................................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................ACTTAAGAAGATCACGATAA............................................................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................ACCGAGACGGAAGATAGGG............................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................GTCGGCTGCGGACACCACGAT................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................AAAACCATAAAGGAGTCTGGTA..................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................CGAGCCGGTAGAGAGGTCGC............................................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................AGAATTGGGGTGGAAAGTT................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group3:5953881-5954131 - | dps_1580 | TCCACTGGGGTTTCTTCGCTGATTCTGGCCTACAGCATCCGACGCTTATGGTGCTAGAGGGCTGCCACTCGGGGCACCTGGCCGAAGCCCTTGAGGTAGCTGCTGAAGGTAGAAATCCACCTGGCTCTGCCTTCTTTCCAGCGCCATAGGGTCTGAATGCCACCTTTCAAATGGATTTTTCTAGTGCTATTAACAGCGTGTACTTTTCTACTCGTATTTCCTCAGACCATAGAATGCCATGGAATACATTT |
| droPer2 | scaffold_1:7441687-7441876 - | ACCC----GGTCTCTTC---------------------------------------------------------CTCCTGGCCGAAGCGCTTGAGGTAGCTGCTGAAGGTAGAAATCCACCTGGCTCTGCCTTCTTTCCAGCGCCATAGGGTCTGAATGCCACCTTTCAAATGGATTTTTCTAGTGCTATTAACAGCGTGTCATTTTCTACTCGTATTTCCTCAGACCAGAGAATGCCATGGAATACATTT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 01:02 AM