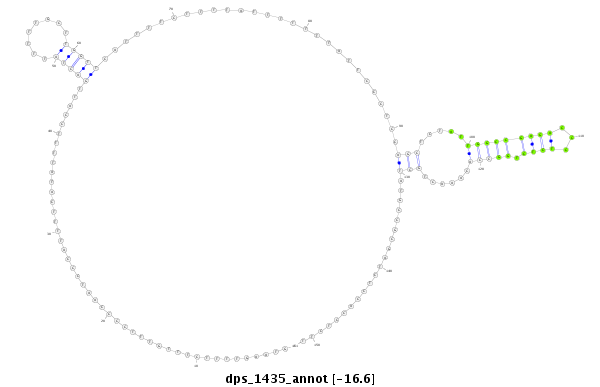
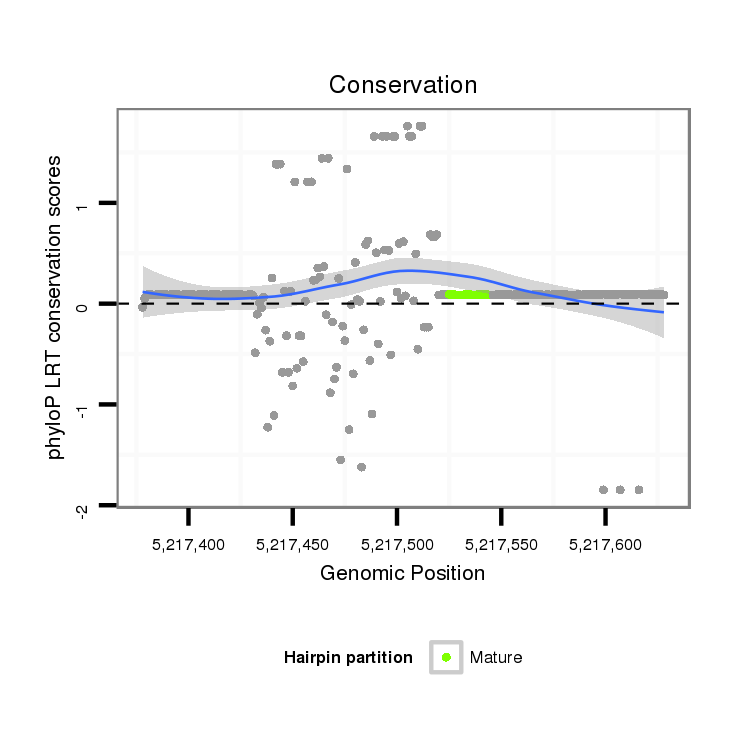
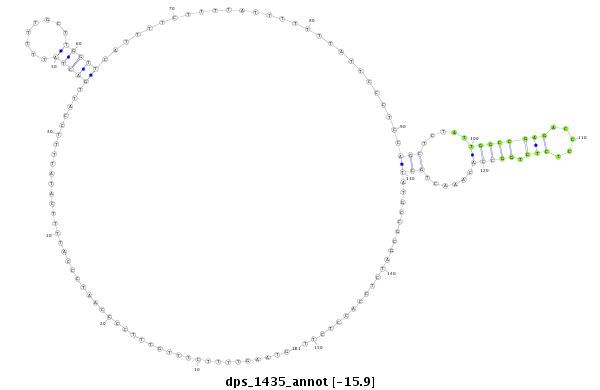
ID:dps_1435 |
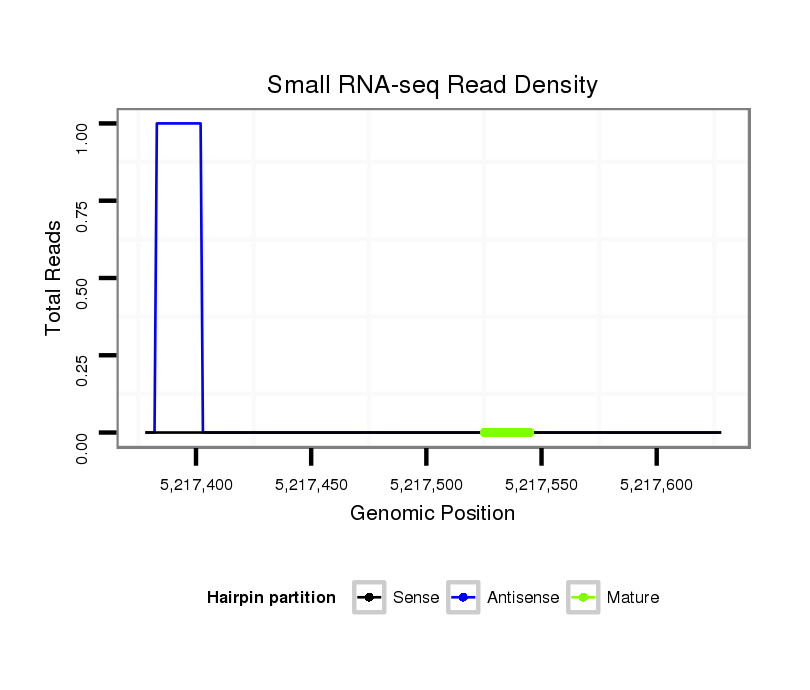
Coordinate:4_group1:5217428-5217578 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.9 | -15.9 | -15.9 |
 |
 |
 |
CDS [Dpse\GA25482-cds]; exon [dpse_GLEANR_17099:1]; intron [Dpse\GA25482-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCGATCGTCTTTCCTTCAGCACAATGCTTTACCTCATCGACTCTGACAGGGTAAGTTTTCTTTGTTTCCCCAATCCCATTTTCATATTTTCCATTGACTATTTTTGCTTGGTTCATTTTCTTTTATTTTTTTATTCCCTCCAGCTCTATTGGCCGAGACCCTCTCTGGCCACAAACTGCTATGCCGCGATCTCCACCTCTTATTCCGCCAGGTAAAGTTATTAAGTTTTCATTTATTACTTACATCATTAC **************************************************.............................................(((((........)))))............................(((.....(((((((((...)))).)))))......))).....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|
| .........................................................................................................GCTGGGCTAATTTTCTTTTATTT........................................................................................................................... | 23 | 3 | 5 | 0.20 | 1 | 0 | 1 |
| .......................................................................................................................................................................................................GTATTCAGCTAGGTAAAGT................................. | 19 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| ...................................................................................................................................................ATTGGCCGAGACCCTCTCTGG................................................................................... | 21 | 0 | 8 | 0.13 | 1 | 1 | 0 |
|
CGCTAGCAGAAAGGAAGTCGTGTTACGAAATGGAGTAGCTGAGACTGTCCCATTCAAAAGAAACAAAGGGGTTAGGGTAAAAGTATAAAAGGTAACTGATAAAAACGAACCAAGTAAAAGAAAATAAAAAAATAAGGGAGGTCGAGATAACCGGCTCTGGGAGAGACCGGTGTTTGACGATACGGCGCTAGAGGTGGAGAATAAGGCGGTCCATTTCAATAATTCAAAAGTAAATAATGAATGTAGTAATG
**************************************************.............................................(((((........)))))............................(((.....(((((((((...)))).)))))......))).....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
GSM343916 embryo |
V112 male body |
M062 head |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....GCAGAAAGGAAGTCGTGTTA.................................................................................................................................................................................................................................. | 20 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................GGGTTAGGGTAAAAGTATAAA.................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....GCAGAAAGGAAGTCGTGTT................................................................................................................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................GTTACGAAATGGAGTAGCTG.................................................................................................................................................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................CGAAATGGAGTAGCTGAGAC.............................................................................................................................................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........AGGAAGTCGTTATACGAAA............................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................AAAAAAATAAGGGAGTTCGT.......................................................................................................... | 20 | 2 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CGAGATAACCGGCTCTGGGAG........................................................................................ | 21 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AAAAAAATAAGGGAGGTCAT.......................................................................................................... | 20 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ACAAAGAGGTTAGAGTAA........................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................ATAAGGGAGGTCGAGATAACCG.................................................................................................. | 22 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GGGTAACAGTATATAAGGTG............................................................................................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................GGAAATAAAAAAATAAAGGA................................................................................................................ | 20 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................CGAAAAAGTATAATAGGTAA............................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................AGGGTTTAGGGTAACAGTAG..................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group1:5217378-5217628 + | dps_1435 | GCGATCGTCTTTCCTTCAGCACAATGCTTTACCTCATCGACTCTGACAGGGTAAGTTTTCTTTGTTT-----CCCCAATCCCATTTTCATATTTTCCATTGACTATTTTTGCTTGGTTCATTTTCTTTTATTTTTTTATTCCCTCCAGCTCTATTGGCCGAGACCCTCTCTGGCCACAAACTGCTATGCCGCGATCTCCACCTCTTATTCCGCCAGGTAAAGTTATTAAGTTTTCATTTATTACTTACATCATTAC |
| droPer2 | scaffold_130:94856-95106 + | GCGATCGTCTTTCCTTCAGCACAATGCTTTACCTCATCGACTCTGACAGGGTAAGTTTTCCTTGTTT-----CCCCAATCCCATTTTCATATTTTCCATTGACTATTTTTGCTTGGTTCATTTTCTTTTATTTTTTTATTCCCTCCAGCTCTATTGGCCGAGACCCTCTCTGGCCACAAACTGCTATGCCGCGATCTCCACCTCTTATTCCGCCAGGTAAAGTTATCAAGTTTTTATTTATTATTTACATCATTAC | |
| droVir3 | scaffold_12928:5177279-5177364 - | GT---------------------------------------------------TT---TTATTATTTGCCTGCCTCATTATTATTTTCTTCTTATTTCTCCTCTATTTTATTCTATTCCATTTTTTTTTTTTTTTTTTTT-------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6500:2034767-2034818 + | TT---------------------------------------------------TTGCTGCAACTTTT-------------------------------------TTCGTTCCTTGATTCATTTTCTTCTTTTTTTTTTTT-------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15252:15942267-15942308 + | TT---------------------------------------------------TT------------------------------------------------------CCCTTGGTTTGTTTTTTTTTTTTTTTATATTTTTTCCA------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000490996:544022-544092 - | TG---------------------------------------------------ACTTCTCTTTTTTT-----GCCGACTCCCCATTTCACATTCTCCTCTCATTTGTTTTGCTT-------------TGGCATTTTGTTT-------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302497:32229-32271 - | TT---------------------------------------------------TT--------------------------------CATATCTT---------ATATTTTCTTTGTTAATTTTTTTTTATTT-----TT-------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000408706:426-505 - | TT---------------------------------------------------TT----TTATATTT-----TTTTTATATTTTTTTTATATTTTTTTATATTTTTTTATATTTTTTTTATATTTTTTTATATTTTTTTT-------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4929:22427828-22427882 + | TT---------------------------------------------------TC----------------------------------TATTTTACATTGAGTCACTCTTATCATTTTGTTTCCTTCTATTTTTATATT-------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 12:43 AM