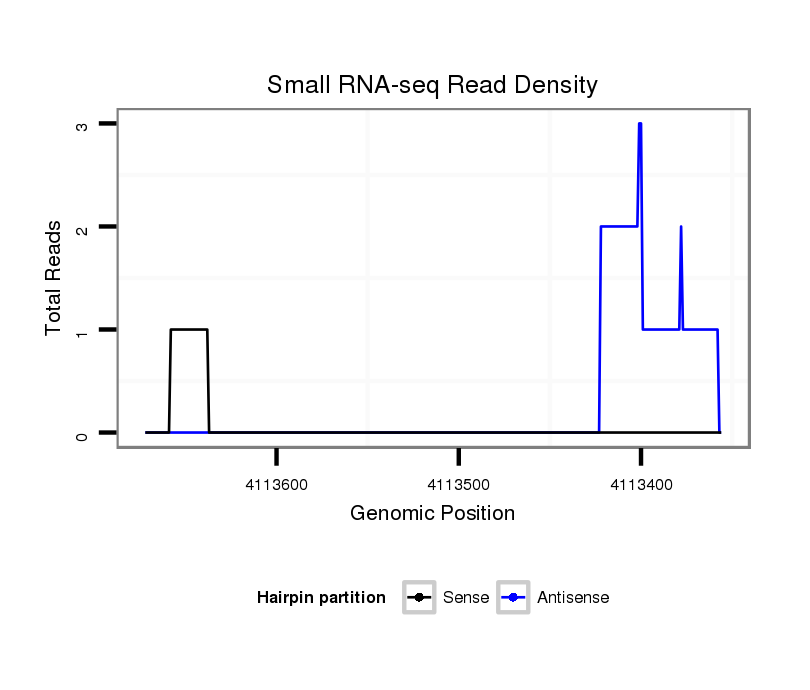
ID:dps_1411 |
Coordinate:4_group1:4113406-4113622 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dpse_GLEANR_16568:3]; CDS [Dpse\GA25198-cds]; CDS [Dpse\GA25198-cds]; exon [dpse_GLEANR_16568:2]; intron [Dpse\GA25198-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCCTGCATCAAGTGCGACGGAAGGGAAGAGCTCTGCCCGTGGCTCAGACAGTGCTACGGAAAAGAAGAAGGGTTCCCCCCGTGGCTCGAATCAGAGTGTTCTATCTATTGCTAATGAAAGGAAGAAGAGTTCCCCCCGTGGCTCGAATCAGAGTGTTCTATCTATTGCTAAGGAAAGGAAGAAAAGTTCCCCCCGTGGCTCGAATCAGAGGGTTCAATCTTCAGGTATTGCTAAGGAAAAGAACAGTTCTCCTCGTGGCTCAGACAGTGCCGTAGCAGTTCGGAGTACAAGATCCTCAGTCAGTGCTACCGAGCGAA **************************************************..(((((((...((((...(((....))).....(((......)))((((((.(((...((.((((....(((((.(((....(((..(((.((((...(((........)))...((((.((...((((......))))..)).)))))))).)))..))))))..)))))...))))))...)))..)))))).))))..)))))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
GSM343916 embryo |
V112 male body |
M040 female body |
|---|---|---|---|---|---|---|---|---|---|
| ..............CGACGGAAGGGAAGAGCTCTG.......................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................ACGGACAAGAAGGAGGGTT................................................................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................AGTGTGTACTATCTATTGC..................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................AGTGTGTACTATCTATTGC.............................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .....................AGGGAAGAGCTCTGCCCGTGG................................................................................................................................................................................................................................................................................... | 21 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 |
| .................CGGAAGGGAAGAGCTCTGCC........................................................................................................................................................................................................................................................................................ | 20 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .....................AGGGAAGAGCTCTGCCCGTGGC.................................................................................................................................................................................................................................................................................. | 22 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CCTTCCGGTATTGCTAGGG................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................................GAACAGTTGTCCTCGTGCA.......................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .......TCAAGTGTGACGGGAGGCA................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
AGGACGTAGTTCACGCTGCCTTCCCTTCTCGAGACGGGCACCGAGTCTGTCACGATGCCTTTTCTTCTTCCCAAGGGGGGCACCGAGCTTAGTCTCACAAGATAGATAACGATTACTTTCCTTCTTCTCAAGGGGGGCACCGAGCTTAGTCTCACAAGATAGATAACGATTCCTTTCCTTCTTTTCAAGGGGGGCACCGAGCTTAGTCTCCCAAGTTAGAAGTCCATAACGATTCCTTTTCTTGTCAAGAGGAGCACCGAGTCTGTCACGGCATCGTCAAGCCTCATGTTCTAGGAGTCAGTCACGATGGCTCGCTT
**************************************************..(((((((...((((...(((....))).....(((......)))((((((.(((...((.((((....(((((.(((....(((..(((.((((...(((........)))...((((.((...((((......))))..)).)))))))).)))..))))))..)))))...))))))...)))..)))))).))))..)))))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
V112 male body |
GSM444067 head |
GSM343916 embryo |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................................................GGAGCACCGAGTCTGTCACGGCA............................................ | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................CATCGTCAAGCCTCATGTTCTAGG...................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................GAGTCAGTCACGATGGCTCGC.. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................TTTCTTGGCATGAGGAGC.............................................................. | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................GATTCAGTCACGGTGGCT..... | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................CAACATAGATAACGATGACTG....................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................CGTCATCGTCAAGCTGCATGT............................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................TTTCTTGGCGAGAGGAGC.............................................................. | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................GGCATCGTCAAGAGGCATGT............................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................GGCATGGTCAAGACTCA............................... | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ..............................................................................................................................................................................................................................................TTCTTGTCATGAAGACCAC............................................................ | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group1:4113356-4113672 - | dps_1411 | TCCTGCATCAAGTGCGACGGAAGGGAAGAGCTCTGCCCGTGGCTCAGACAGTGCTACGGAAAAGAAGAAGGGTTCCCCCCGTGGCTCGAATCAGAGTGTTCTATCTATTGCTAATGAAAGGAAGAAGAGTTCCCCCCGTGGCTCGAATCAGAGTGTTCTATC------TATTGCTAAGGAAAGGAAGAAAAGTTCCCCCCGTGGCTCGAATCAGAGGGTTCAATCTTCAGGTATTGCTAAGGAAAAGAACAGTTCTCCTCGTGGCTCAGACAGTGCCGTAGCAGTTCGGAGTACAAGATCCTCAGTCAGTGCTACCGAGCGAA |
| droPer2 | scaffold_5:1150621-1150943 + | ACCTGCATCAAGTGCAACGGAAGGGAAGAGCTCTGCCCGTGGCTCAGACAGTGCTACGGAAAAGAAGAAGAGTTCTCCAAGGGGCTCGAATCAGAGTGCTCTATCTATTGCTAAGCAAAGGAAGAAGAGCTCTCCCAGTGGCTCGAATCAGGCTATCCTATCTTCAGGTATTTCTAAGGAAAGGAAGAAGAGTTCCCCCCGTGGCTCGAATCACAGTGTCCAATCTTCACGTATTTCTAAGGAAAAGAACAGTTCTCCTCGTTGCTCAGACGGTGGCAGAGCAATTCGGAGTCCAAGATCTTCAGTCAGCGCTAGCGAACGAA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 12:39 AM