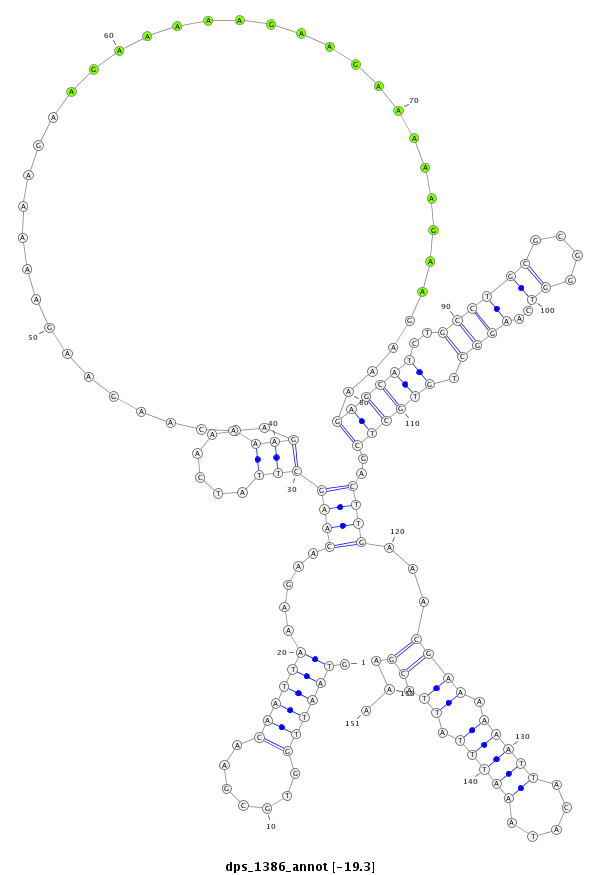
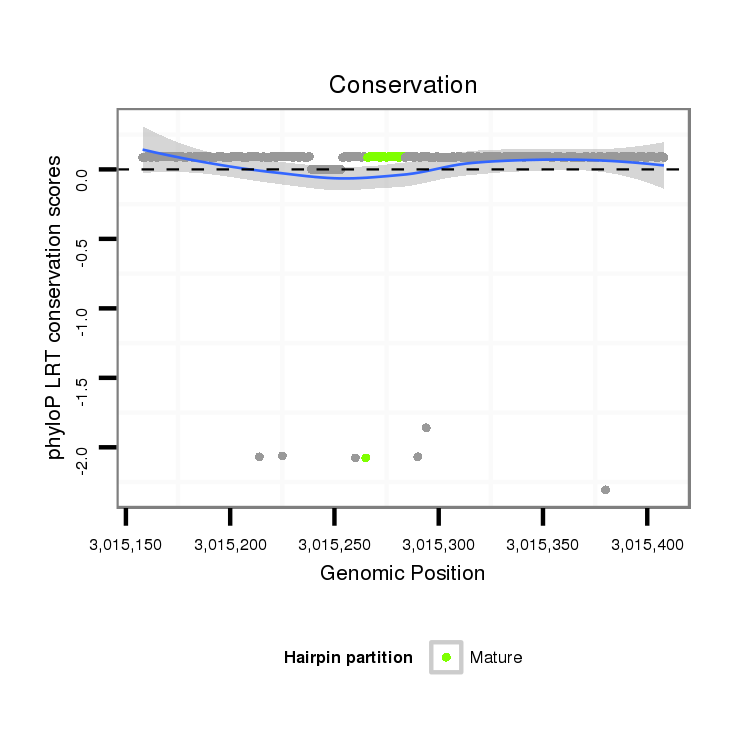
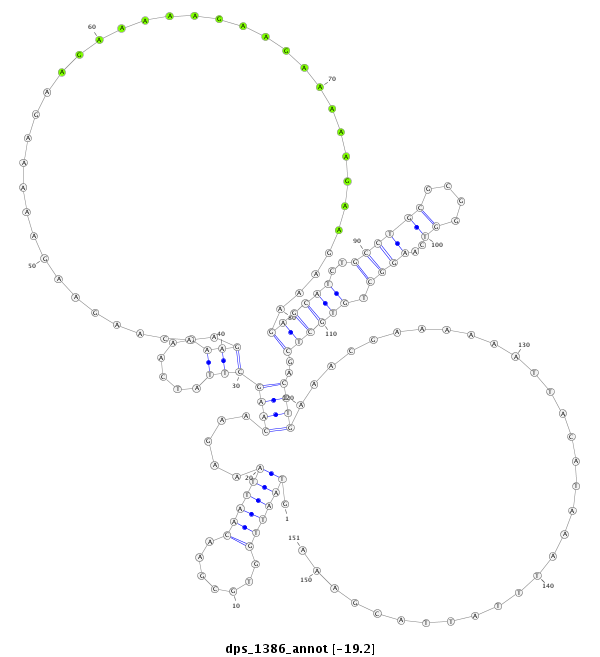
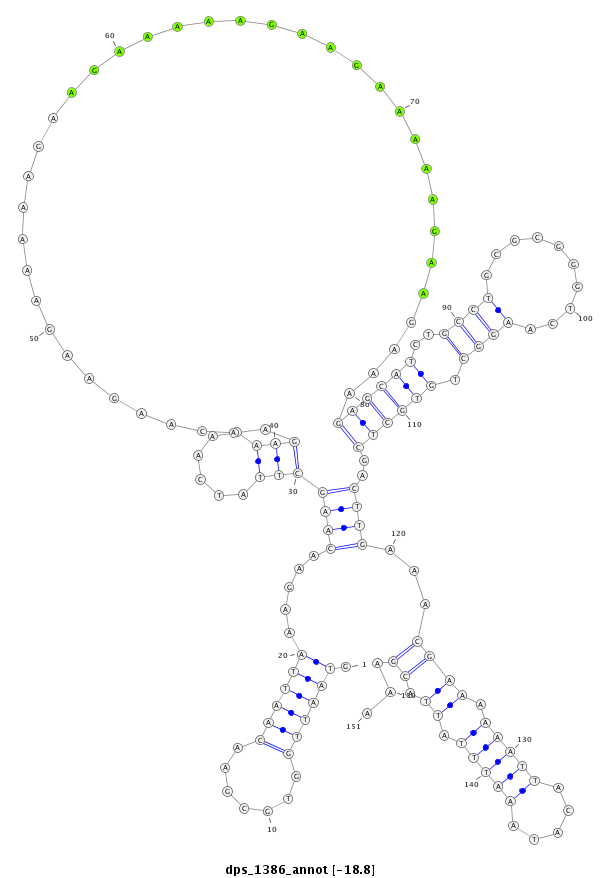
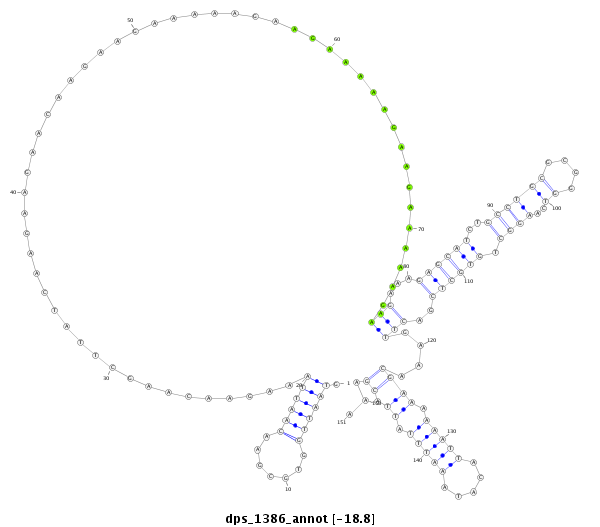
ID:dps_1386 |
Coordinate:4_group1:3015208-3015358 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.2 | -18.8 | -18.8 |
 |
 |
 |
exon [dpse_GLEANR_16982:1]; CDS [Dpse\GA25422-cds]; intron [Dpse\GA25422-in]
| Name | Class | Family | Strand |
| (GAA)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCATCAAGAATTAAAACTTTGGAAGTCTATAATCCGTAACCCGATTAACCGTAATTGGTGCGAACAATTAAAGAACAAGCTTATCAAGAAGAACAAGAAGAAAAAGAAGAAAAAGAAGAAAAAGAAGAAAGAGCATCTGCCTGCGCGGGTCAAGGCTGTGCTCGACTTGAAACGAAAAAATTACATAAATTTATTACGAAAGCCAACAGGCCATGGTGCAAGGAAATACCTGAAAAGGTTTGGTAAGGATC **************************************************.((((((.......)))))).....(((((((......))).......................................((((((..((((((....))..)))).))))))..))))...((((.(((((.....))))).)).))...************************************************** |
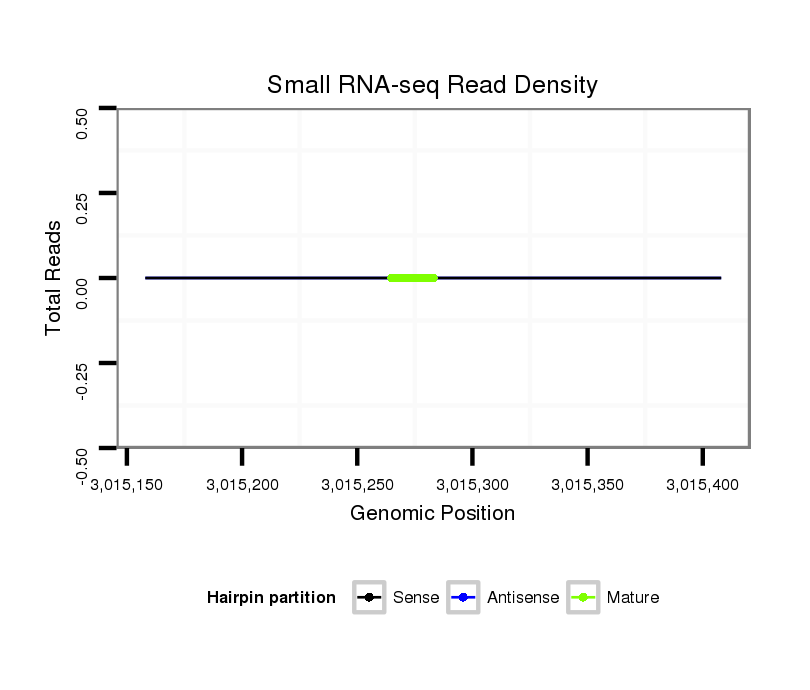
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
SRR902011 testis |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................CAAGAAGACAACGAAGAAAAAGCAG..................................................................................................................................... | 25 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 |
| ......................................................................................AGAAGAGCAAGAAGAAAAAG................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................TGGTGCGAACAATTAAAGAAA............................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................GGATATACCTGAGAAGGAT........... | 19 | 3 | 18 | 0.39 | 7 | 5 | 0 | 0 | 2 |
| ...........................................................................................................AGAAAAAGAAGAAAAAGAA............................................................................................................................. | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................AGAAAAAGAAGAAAAAGAA...................................................................................................................................... | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................AAGAAGAAAAAGAAGAAAAGG............................................................................................................................... | 21 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................AAGAAGAAAAAGAAGAAAAGG........................................................................................................................................ | 21 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................AGAAGAGCAAGAAGAAAAA.................................................................................................................................................. | 19 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................AGAAGAACAAGAAGAGATTGAAG.............................................................................................................................................. | 23 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................AAGGAAGAAAAAGAAGAAAA................................................................................................................................. | 20 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................CAAGCTCATGAAGAAGAAC............................................................................................................................................................. | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................AGAAGAGAAGGAAGAATAAGAAGA.................................................................................................................................... | 24 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................AGAAGAGAAGGAAGAATAAGAAGA........................................................................................................................... | 24 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................AAAGAAGAAATAGAAGAA................................................................................................................................... | 18 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................AAAGAAGAAATAGAAGAA.......................................................................................................................... | 18 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GAAATACGTGAAAAGATTT.......... | 19 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................AAAAAGAAGAAAAAGAAGGA.......................................................................................................................... | 20 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................AAAAAGAAGAAAAAGAAGGA................................................................................................................................... | 20 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
|
CGTAGTTCTTAATTTTGAAACCTTCAGATATTAGGCATTGGGCTAATTGGCATTAACCACGCTTGTTAATTTCTTGTTCGAATAGTTCTTCTTGTTCTTCTTTTTCTTCTTTTTCTTCTTTTTCTTCTTTCTCGTAGACGGACGCGCCCAGTTCCGACACGAGCTGAACTTTGCTTTTTTAATGTATTTAAATAATGCTTTCGGTTGTCCGGTACCACGTTCCTTTATGGACTTTTCCAAACCATTCCTAG
**************************************************.((((((.......)))))).....(((((((......))).......................................((((((..((((((....))..)))).))))))..))))...((((.(((((.....))))).)).))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
V112 male body |
|---|---|---|---|---|---|---|---|---|
| .......................................................................................CTTCTTGTTCGTCTGTTTCTT............................................................................................................................................... | 21 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................CGCCCAGTTGCGACTCGGGCT...................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ......................................................................................TCTTTTTGTTCTTCTTTTTCTTGT............................................................................................................................................. | 24 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ........................................................................................TTCTTGTTCGTCTGTTTCTT............................................................................................................................................... | 20 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................CCCTTTTGGTTGTCCGGTA..................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group1:3015158-3015408 + | dps_1386 | GCATCAAGAATTAAAACTTTGGAAGTCTATAATCCGTAACCCGATTAACCGTAATTGGTGCGAACAATTAAAGAACAAGCTTATCAAGAAGAACAAGAAGAAAAAGAAGAAAAAGAAGAAAAAGAAGAAAGAGCATCTGCCTGCGCGGGTCAAGGCTGTGCTCGACTTGAAACGAAAAAATTACATAAATTTATTACGAAAGCCAACAGGCCATGGTGCAAGGAAATACCTGAAAAGGTTTGGTAAGGATC |
| droPer2 | scaffold_5:2205142-2205377 - | GCATCAAGAATTAAAACTTTGGAAGTCTATAATCCGTAACCCGATTAACCGTAATTTGTGCGAACAAGTAAAGAACAAGCT---------------GAAGAACAAGACGAAAAAGAAGAAAAAGAAGAAAGATCATTTGCCTGCGCGGGTCAAGGCTGTGCTCGACTTGAAACGAAAAAATTACATAAATTTATTACGAAAGCCAACAGGCCATGGTGCAAGCAAATACCTGAAAAGGTTTGGTAAGGATC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 12:37 AM