
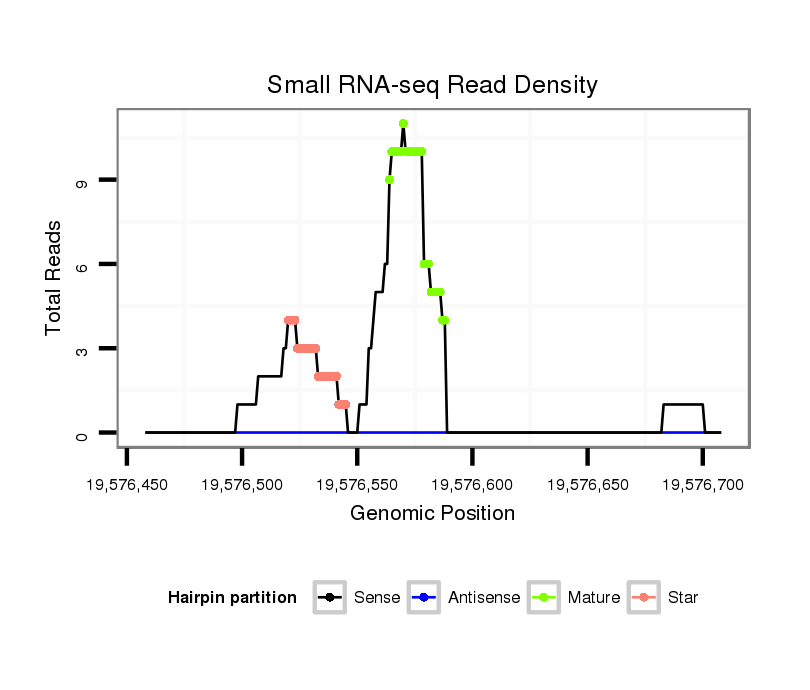
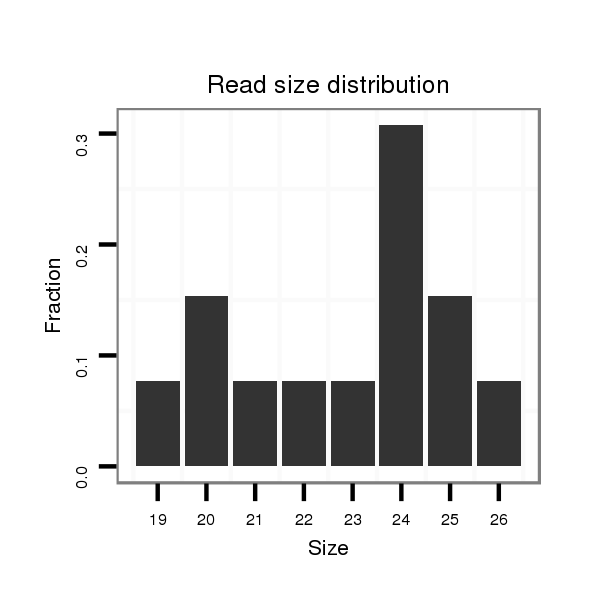


ID:dps_1325 |
Coordinate:3:19576508-19576658 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.8 | -16.4 | -16.4 |
 |
 |
 |
exon [dpse_GLEANR_16460:2]; CDS [Dpse\GA25144-cds]; intron [Dpse\GA25144-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCCGAAGCTATAGGCTCGATCAGTTGCAGCTCATTTCAAACCTCGCCGGCGACATGATCGGCTGTGTGTCGGGGAGCTTGACGTTTTGCCTGCTGGCAGCACTTATCGGAAGCGGACTGGGAGCAGAGGACTGTTAGCAGAGGTCTCGGAGCATCGCTGATAGGCGGGCACTAATTGCGAAATATTTTACGATTGTTTTAGATCCCGACTATTGTAGCACCTTTGAGGGGTGGCCTGGGTGGTGCGTCCAC **************************************************************..(((.((.((..((((((.((.(((((....))))).))...))..))))...)).)).)))......************************************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902012 CNS imaginal disc |
M040 female body |
SRR902011 testis |
M022 male body |
GSM444067 head |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................CGGAAGCGGACTGGGAGCAGAGGAC........................................................................................................................ | 25 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .................................................................................................AGCACTTATCGGAAGCGGACTGGG.................................................................................................................................. | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATCGGAAGCGGACTGGGAGC............................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................CCTCGCCGGCGACATGATCGGCTGTG......................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TGTGTGTCGGGGAGCTTGACGTTTTG................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................CGGACTGGGAGCAGAGGAC........................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................CACTTATCGGAAGCGGACTGGG.................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GCTGTGTGTCGGGGAGCTTGACGT....................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................CGACATGATCGGCTGTGTGTCGGGGA................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................AGGGGTGGCCTGGGTGGT........ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................GGAAGCGGACTGGGAGCAGAGGAC........................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TGGCAGCACTTATCGGAAGC.......................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CGGAAGCGGACTGGGAGCAGAGG.......................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................ACTTATCGGAAGCGGACTGGG.................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGCTAGCAGCTCTTACCGG.............................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CAGGTTTGAGGGGTGGTCTG.............. | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................TCGGGTAGCTAGAGGTTTT.................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................GTTTGTAGAGGGCTCGGAGC................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CGGCTTCGATATCCGAGCTAGTCAACGTCGAGTAAAGTTTGGAGCGGCCGCTGTACTAGCCGACACACAGCCCCTCGAACTGCAAAACGGACGACCGTCGTGAATAGCCTTCGCCTGACCCTCGTCTCCTGACAATCGTCTCCAGAGCCTCGTAGCGACTATCCGCCCGTGATTAACGCTTTATAAAATGCTAACAAAATCTAGGGCTGATAACATCGTGGAAACTCCCCACCGGACCCACCACGCAGGTG
************************************************************************************************************************..(((.((.((..((((((.((.(((((....))))).))...))..))))...)).)).)))......************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902012 CNS imaginal disc |
M040 female body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................CGGAATCTAGGGCTGAT........................................ | 17 | 2 | 3 | 1.00 | 3 | 3 | 0 | 0 |
| .......................................................................................................................................................................................TACAATGCTAACCAAACCTA................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 |
| ......................................................................................ACCGACGACCGTAGTGAAAAG................................................................................................................................................ | 21 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| .....................................TTTGGACCGGCCGCCGCACTA................................................................................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................CGGAATCTAGGGCTGATT....................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 3:19576458-19576708 + | dps_1325 | GCCGAAGCTATAGGCTCGATCAGTTGCAGCTCATTTCAAACCTCGCCGGCGACATGATCGGCTGTGTGTCGGGGAGCTTGACGTTTTGCCTGCTGGCAGCACTTATCGGAAGCGGACTGGGAGCAGAGGACTGTTAGCAGAGGTCTCGGAGCATCGCTGATAGGCGGGCACTAATTGCGAAATATTTTACGATTGTTTTAGATCCCGACTATTGTAGCACCTTTGAGGGGTGGCCTGGGTGGTGCGTCCAC |
| droPer2 | scaffold_37:578071-578321 + | GCCGAAGCTATAGGCTCGATCAGTTGCAGCTCATTTCAAACCTCGCCGGCGACATGATCGGCTGTGTGTCGGGGAGCTTGACGTTTTGCCTGCTGGCAGCACTTATCGGAAGCGGACTGGGAGCAGAGGACTGTTAGCAGAGGTCTCGGAGCATCGCTGATAGGCGGGCACTAATTGCGAAATATTTTACGATTGTTACAGATCCCGACTATTGTAGCACCTTTGAGGGGTGGCCTGGGTGGTGCGTCCAC | |
| droKik1 | scf7180000302682:1755104-1755162 + | GTTCTACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCCACAATACTGCACTACCTATGATGAGAAGCCAGGCAGGTGTCTTCGC | |
| droFic1 | scf7180000453809:79063-79114 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATCCCACCTACTGCACCACCTTTGAAGAGAAACCAGGGAAATGCATACAC | |
| droEug1 | scf7180000409672:479825-479876 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATCCCGAATACTGCACCACCTTCGATAATAAACCAGGTAAATGTGTAGAC | |
| droSim2 | 2r:12253265-12253316 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATCCAACATACTGCACCACCTACGATCAGAAGCCAGGCAAATGCGTACAC | |
| droSec2 | scaffold_1:9080477-9080528 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATCCAACATACTGCACCACCTACGATCAGAAGCCAGGCAAATGCGTACAC | |
| droEre2 | scaffold_4845:14278514-14278565 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATCCCACATACTGCACTACGTACGATCAGAAGCCAGGCAAATGCGTAAAC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 12:26 AM