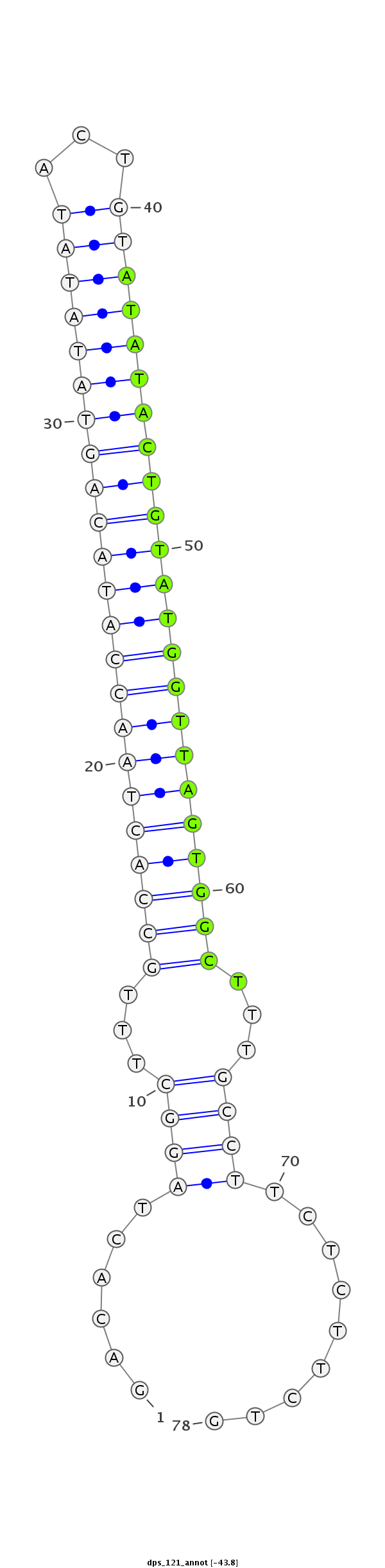
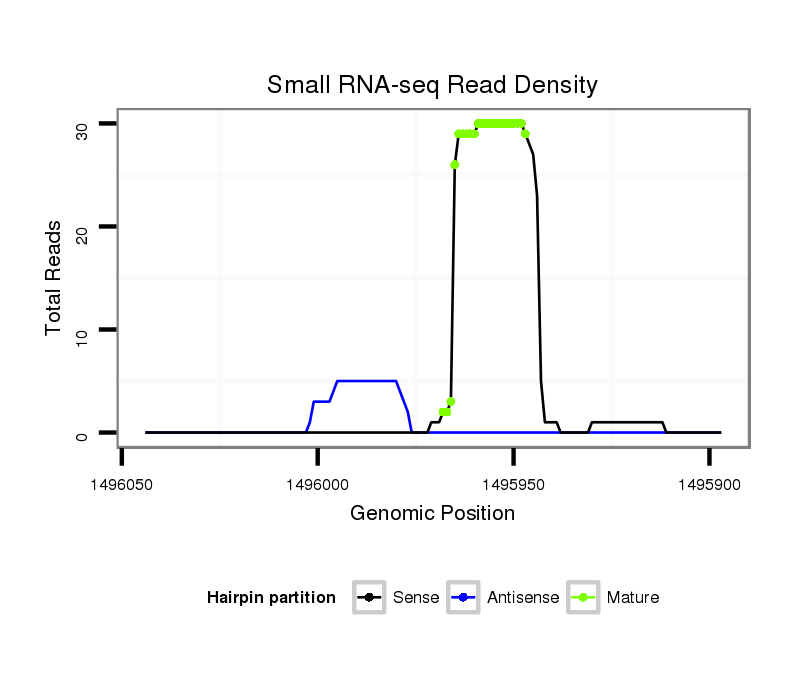
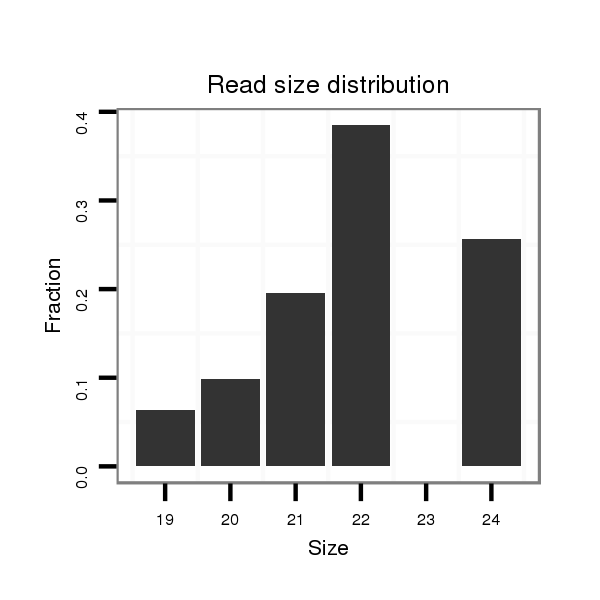
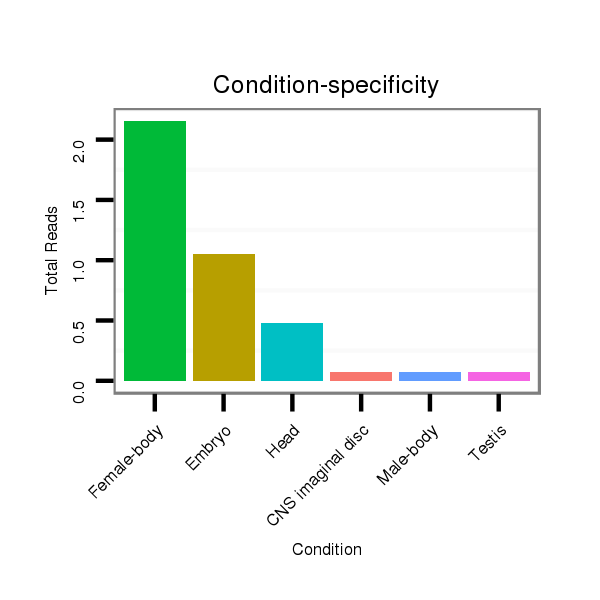
ID:dps_121 |
Coordinate:4_group4:1495947-1495994 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -42.9 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
GGGAAAATTTGTTTTTGAAGCCCTTACGGTGGTACGACACTAGGCTTTGCCACTAACCATACAGTATATATACTGTATATACTGTATGGTTAGTGGCTTTGCCTTCTCTTCTGTGCTGTGAAGAGGGAGAGAGAGATAGAATGCGAAC
***********************************......((((...(((((((((((((((((((((((...)))))))))))))))))))))))...)))).........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
V112 male body |
M059 embryo |
GSM444067 head |
SRR902011 testis |
M062 head |
SRR902010 ovaries |
SRR902012 CNS imaginal disc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................TACTGTATGGTTAGTGGCTTTG............................................... | 22 | 0 | 1 | 16.00 | 16 | 11 | 1 | 2 | 0 | 0 | 1 | 0 | 1 | 0 |
| ...............................................................................TACTGTATGGTTAGTGGCTTT................................................ | 21 | 0 | 1 | 4.00 | 4 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TACTGTATGGTTAGTGGCTTTGC.............................................. | 23 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................AAGCCATTACGCCGGTACGA............................................................................................................... | 20 | 3 | 6 | 2.33 | 14 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................ACTGTATGGTTAGTGGCTTTG............................................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATATACTGTATGGTTAGTGGCT.................................................. | 22 | 0 | 2 | 1.50 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGTATATACTGTATGGTTAGTGGC................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................ATGGTTAGTGGCTTTGCCTTC.......................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGTATATACTGTATGGTTAGTGACTT................................................. | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................GCTGTGAAGAGGGAGAGAG............... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TACTGTATGGTTAGTGGCTTTT............................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................ATACTGTATGGTTAGTGGCTT................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................ACTGTATGGTTAGTGGCTTTGC.............................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................ATACTGTATGGTTAGTGGCTTTT............................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TACTGTATGGTTAGTGGCTTTTCCT............................................ | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................ACTGTATGGTTAGTGGCTTTTCCT............................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............TTAAGCCATTACGCTGGTACG................................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TTGCCACTAACCATACAGTATA................................................................................ | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATATACTGTATGGTTAGTGGC................................................... | 21 | 0 | 14 | 0.43 | 6 | 1 | 0 | 1 | 0 | 2 | 1 | 0 | 0 | 1 |
| ..............................................................................ATACTGTATGGTTAGTGGCT.................................................. | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATATACTGTATGGTTAGTGGCTGT................................................ | 24 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATATACTGTATGGTTAGTGACTTT................................................ | 24 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TATACTGTATGGTTAGTGGCT.................................................. | 21 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CTGTGTAGAGGGATAGAGA.............. | 19 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TATACTGTATGGTTAGTGACTT................................................. | 22 | 1 | 8 | 0.25 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................ATACTGTATGGTTAGTGGC................................................... | 19 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TGGGATGTGAAGAGGGACAGAG............... | 22 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......ATTTGTTTTCTAAGCCCTCAC......................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATATACTGTATGGTTAGTGGCTG................................................. | 23 | 1 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TACTGTATGTACTGTATGTTTAGTGT.................................................... | 26 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................TGGGCTGTAAAGAGGGAGGGAG............... | 22 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GTATACTGTATGGTTAGTGGCTT................................................. | 23 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AACCGTTACGGTGGTACGT............................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GTATACTGTATGGTTAGTGGCTTG................................................ | 24 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TATACTGTATGGTTAGTGACTTC................................................ | 23 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GTATATACTGTATGGTTACTG..................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GGATATACTGTATGGTTC........................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................TTTATACTGTATGGTTAGTGGC................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATATACTGTATGGTTAGTGACTTTA............................................... | 25 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TATACTGTATGGTTAGTGGC................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CCCTTTTAAACAAAAACTTCGGGAATGCCACCATGCTGTGATCCGAAACGGTGATTGGTATGTCATATATATGACATATATGACATACCAATCACCGAAACGGAAGAGAAGACACGACACTTCTCCCTCTCTCTCTATCTTACGCTTG
***********************************......((((...(((((((((((((((((((((((...)))))))))))))))))))))))...)))).........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M059 embryo |
GSM444067 head |
SRR902010 ovaries |
SRR902011 testis |
SRR902012 CNS imaginal disc |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................GATCCGAAGCAGTGACTGG.......................................................................................... | 19 | 3 | 20 | 2.55 | 51 | 46 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTGATTGGTATGTCATAT................................................................................ | 20 | 0 | 20 | 1.95 | 39 | 37 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| .................................................GGTGATTGGTATGTCATAT................................................................................ | 19 | 0 | 20 | 1.65 | 33 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................CCGAAACGGTGATTGGTATGTCAT.................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CGAAACGGTGATTGGTATGTCATA................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CGAAACGGTGATTGGTATGTCA................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................ACGGTGATTGGTATGTCATAT................................................................................ | 21 | 0 | 17 | 0.65 | 11 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AACGGTGATTGGTATGTCAT.................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTGATTGGTATGTCATATA............................................................................... | 21 | 0 | 14 | 0.43 | 6 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 0 |
| ..............................................AACGGTGATTGGTATGTCATATG............................................................................... | 23 | 1 | 16 | 0.38 | 6 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .................TTCGGTAATGCGGCCATGCT............................................................................................................... | 20 | 3 | 6 | 0.33 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTGATTGGTATGTCATA................................................................................. | 19 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AGATCCGAAGCAGTGATTG........................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGATTGGTATGTCATATA............................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTGATTGGTATGTCATATGTA............................................................................. | 23 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................AAACGGTGATTGGTATGTCATATG............................................................................... | 24 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................ATCCGAAGCAGTGACTGGT......................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AAAGGTGATTGGTATGTCAT.................................................................................. | 20 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTGATTGGTATGTCATA................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTGATTGGTATGTCATATTT.............................................................................. | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................ACTGTGATTGGTATGTCATAT................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 4_group4:1495897-1496044 - | dps_121 | GGGAAAATTTGTTTTTGAAGCCCTTACGGTGGTACGACACTAGGCTTTGCCACTAACCATACAGTATATATACTGTATATACTGTATGGTTAGTGGCTTTGCCTTCTCTTCTGTGCTGTGAAGAGGGAGAGAGAGATAGAATGCGAAC |
| droPer2 | scaffold_10:498026-498116 - | GGGAACATTTGTTTTTGAAGCCCTTACGGTGGTACGACACTAGGCTTTGC---------------------------------------------------------CTTCTTTGCTGTGAAGAGGGAGAGAGAGATAGAATGCGAAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/15/2015 at 02:37 PM