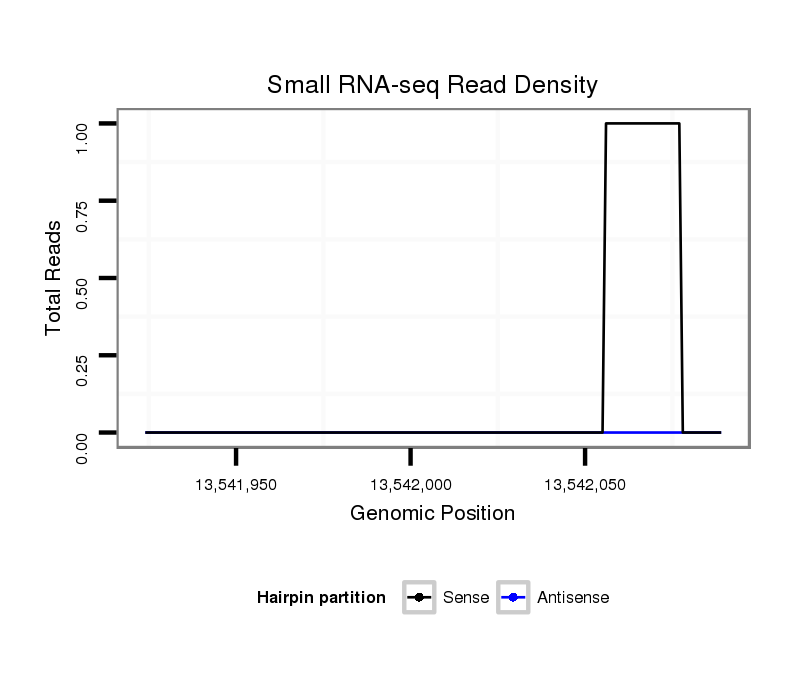
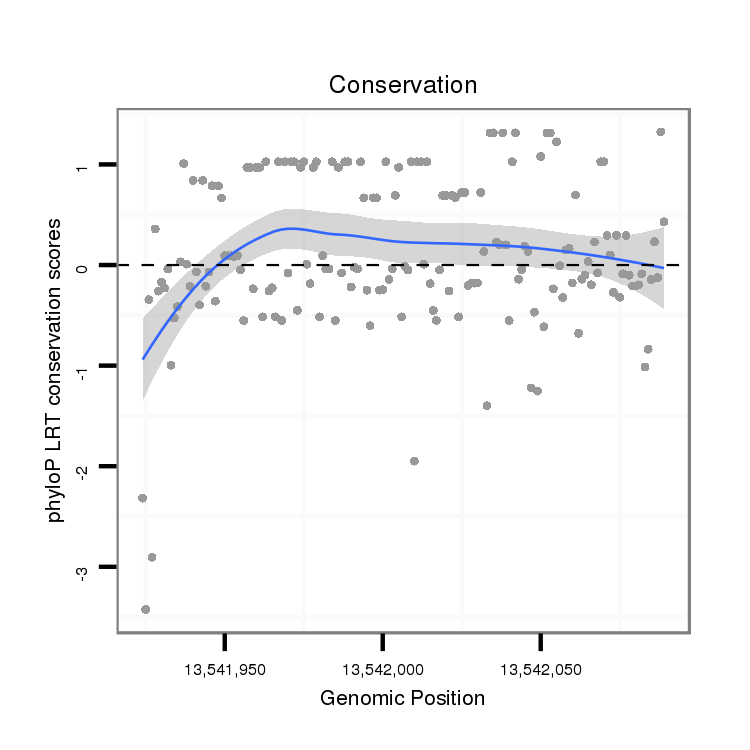
ID:dps_1204 |
Coordinate:3:13541974-13542039 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
intron [Dpse\GA24967-in]; CDS [Dpse\GA24967-cds]; exon [dpse_GLEANR_16041:7]; exon [dpse_GLEANR_16041:6]; CDS [Dpse\GA24967-cds]; intron [Dpse\GA24967-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------################################################-- CCTGAACCATCCGAAGAGCAGTCTGAAAACATTCCGCCTAAGAAGACAACGTGGGTATATTTGCTTATAATGCAAGATTTTCATTTGAACAGGCATTATCTTTATTGTAATTTCAGAAAGCCGTCTTCTTTGAGCTCCGCTCGTGATGACGTAGTAACTATTGGGT **************************************************.........((...((((((((.(((((...................)))))))))))))....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................................CATTTGAAAAGGCAGTATCTA................................................................ | 21 | 3 | 4 | 35.50 | 142 | 112 | 30 | 0 | 0 |
| .................................................................................CATTTGAAAAGGCAGTATCT................................................................. | 20 | 2 | 1 | 32.00 | 32 | 27 | 5 | 0 | 0 |
| ..................................................................................ATTTGAAAAGGCAGTATCT................................................................. | 19 | 2 | 1 | 25.00 | 25 | 24 | 1 | 0 | 0 |
| ................................................................................CCATTTGAAAAGGCAGTATCT................................................................. | 21 | 3 | 2 | 6.50 | 13 | 10 | 3 | 0 | 0 |
| .................................................................................CATTTGAAAAGGCAGTATC.................................................................. | 19 | 2 | 2 | 4.00 | 8 | 7 | 1 | 0 | 0 |
| ..................................................................................ATTTGAAAAGGCAGTATCTA................................................................ | 20 | 3 | 8 | 3.38 | 27 | 27 | 0 | 0 | 0 |
| .................................................................................CATTTGAAAAGGCAGTATCTT................................................................ | 21 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .................................................................................CATTTGAAAAGGCAGTATCTTTT.............................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................TCCGCCTAAGAAGACAACAAAG................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................AGCTCCGCTCGTGATGACGTAG............ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ................................................................................CCATTTGAAAAGGCAGTATC.................................................................. | 20 | 3 | 7 | 0.86 | 6 | 4 | 2 | 0 | 0 |
| ..................................................................................ATTTGAAAAGGCAGTATC.................................................................. | 18 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................................TTTGAAAAGGCAGTATCT................................................................. | 18 | 2 | 8 | 0.38 | 3 | 3 | 0 | 0 | 0 |
| .................................................................................CATTTGAAAAGGCAGTAT................................................................... | 18 | 2 | 9 | 0.33 | 3 | 3 | 0 | 0 | 0 |
| ................................................................................GCATTTGAAAAGGCAGTATCT................................................................. | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................CATTTGAAGAGGCAGTATCTA................................................................ | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................ATTTGAAAAGGCATTATCTAG............................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................TTTGAAAAGGCATTATCTAGA.............................................................. | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................CATTTGAAAAGGCAGTATCA................................................................. | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................ATTTGAAAAGGCAGTATCTG................................................................ | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
|
GGACTTGGTAGGCTTCTCGTCAGACTTTTGTAAGGCGGATTCTTCTGTTGCACCCATATAAACGAATATTACGTTCTAAAAGTAAACTTGTCCGTAATAGAAATAACATTAAAGTCTTTCGGCAGAAGAAACTCGAGGCGAGCACTACTGCATCATTGATAACCCA
**************************************************.........((...((((((((.(((((...................)))))))))))))....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
|---|---|---|---|---|---|---|---|
| ................................................................................GGTAAACTTTTCCGTCATAGA................................................................. | 21 | 3 | 2 | 27.00 | 54 | 54 | 0 |
| .................................................................................GTAAACTTTTCCGTCATAGAT................................................................ | 21 | 3 | 4 | 15.00 | 60 | 59 | 1 |
| .................................................................................GTAAACTTTTCCGTCATAGA................................................................. | 20 | 2 | 1 | 3.00 | 3 | 3 | 0 |
| ..................................................................................TAAACTTTTCCGTCATAGA................................................................. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ................................................................................GGTAAACTTTTCCGTCATAG.................................................................. | 20 | 3 | 7 | 0.86 | 6 | 6 | 0 |
| ..................................................................................TAAACTTTTCCGTCATAGAT................................................................ | 20 | 3 | 8 | 0.38 | 3 | 3 | 0 |
| .................................................................TTATTAGGTTCTGAAAGTAAA................................................................................ | 21 | 3 | 16 | 0.06 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 3:13541924-13542089 + | dps_1204 | CCTGAACCATCCGAAGAGCAGTCTGAAAACATTCCGCCTAAGAAGACAACGTGGGTATATTTGCTTATAATGCAAGATTTTCATTTGAACAGGCATTA-TCT-----TTATTGTAATTTCAGAAA-GCCGTCTTCTTTGAGCTCCGCTCGT-------GATGACGTAGTAA---CTATTGGGT |
| droPer2 | scaffold_2:8077498-8077663 - | CCTGAACCATCCGAAGAGCAGTCTGAAAACATTCCGCCTAAGAAGACAACGTGGGTATATTTGCTTATAATGCAAGATTTTCATTTTAACAGGCATTA-TCT-----TTATTGTAATTTCAGAAA-GCCGTCTTCTTTGAGCTCCGCTCGT-------GATGACGTAGTAA---CTATTGGGT | |
| droWil2 | scf2_1100000004585:351397-351539 - | TCTTTCCGTCG--------------------TGCCCCCAATATATAAAAAGTAAGTT-ATTGGATTTAAA-------TGTTCGCTTAAATAATAATTTGTCATAATT-------AATTACAGCAAGAACATGATATTAGATAG--TTGCACGACGGATTATAAAAGGAAAA---CCCCCGAGT | |
| droMoj3 | scaffold_6496:20560094-20560096 + | GGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droAna3 | scaffold_13266:150199-150322 + | GGAAAAGAG----------------------AACCGCCCA--GAAACAAAGTGAGTG-GCTAGCTTTTGATAGAAAGTTC-CTTATGAACAATTT------CTACTT-------CCTTTCAGTAA---TAATA-ATTTGGACT------AT-------GATGTCGTGGAGG---ACTTAGGGT | |
| droFic1 | scf7180000453201:4495-4518 - | CCAA--CGAACTGAGGAAAAGCCTGA------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000491201:1550832-1550857 - | CAAGAGGGACATGAAAAACAACCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------A | |
| droTak1 | scf7180000415763:178650-178675 + | TGAT-------------------------------------------------------------------------------------------------------------------------------------------------------------GTTGAAGTAATCGTTGGAAGGT | |
| droSim2 | 2r:6558706-6558779 + | ATTT-------------------------------------------------------------------------------------------------------TTTCCATAGTTTTAA--A-ACCATTATTTTAGGACTTCCTTTGTGACAGAT---GTTGAAGTGGACCTTAAAAAGT | |
| droSec2 | scaffold_1:3392157-3392230 + | ATTG-------------------------------------------------------------------------------------------------------TTTCCATAATTTTAA--A-ACCACTTTTTTAGGACTTCCTTTGTGACAGAT---GTTGAAGTGGACCTTAAAAAGT | |
| droYak3 | 2L:18408076-18408082 + | TGAGAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4929:8586335-8586349 - | CCCAAACGATCCAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 12:11 AM