


ID:dps_118 |
Coordinate:Unknown_group_117:19676-19749 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
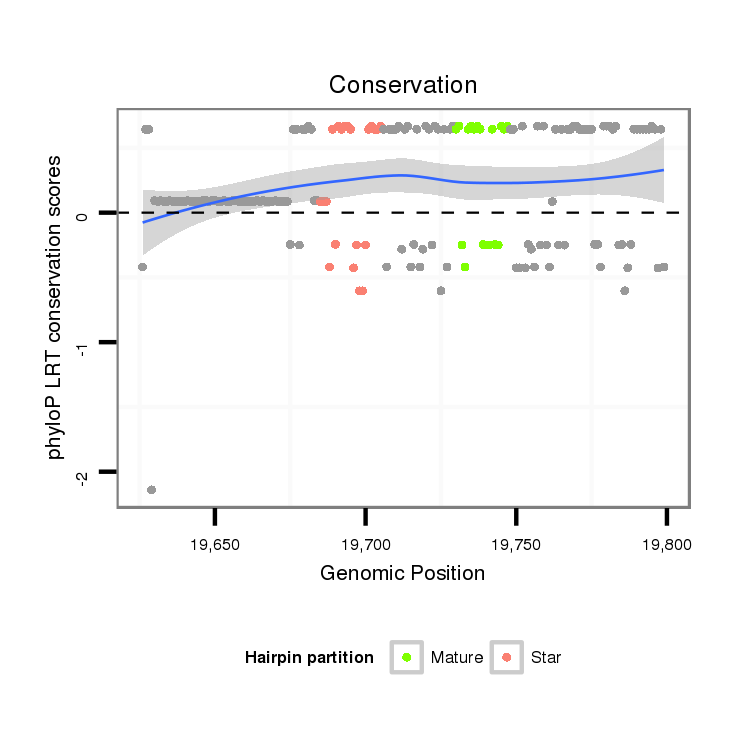
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -40.3 |
 |
intergenic
| Name | Class | Family | Strand |
| Gypsy8-I_Dpse-int | LTR | Gypsy | + |
| mature | star |
|
AGCTTCAGGACGCGCAGGGCAACTACGAGCTTCGGGCCAGACCGACGGGTGCCGCTCATGCGTGTACTACCGGGCGAGGAGTCGGATGATTCTTAGGTCCGAGACACTGTGTGAGGGAGACACCCGTGAAATGAGTCCGGCCGAAGGCGCATAAAAGTGTCCTGGGCCGTGGGT
********************************************(((((((((.(((((((((((.((..(((((.(((((((....)))))))..))))))))))).)))))))))...)))))))..........************************************* |
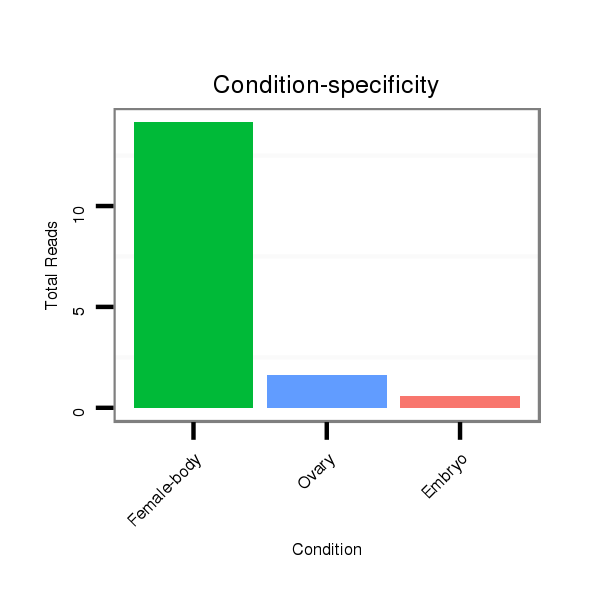
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
SRR902011 testis |
M022 male body |
V112 male body |
GSM343916 embryo |
M059 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................CACTGTGTGAGGGAGACACCCGT............................................... | 23 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACA.................................................... | 18 | 0 | 5 | 4.80 | 24 | 15 | 7 | 0 | 0 | 0 | 2 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACAA................................................... | 19 | 1 | 7 | 3.29 | 23 | 21 | 0 | 0 | 0 | 0 | 1 | 1 |
| ........................................................................................................CACTGTGTGAGGGAGACAC................................................... | 19 | 0 | 5 | 3.20 | 16 | 15 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................CACTGTGTGAGGGAGACACCCG................................................ | 22 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACACC.................................................. | 20 | 0 | 5 | 2.60 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACACCCGTGA............................................. | 25 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCGTGTACTACCGGGCGAGGA.............................................................................................. | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TCGGATGGTTCTTCGGTCAGA........................................................................ | 21 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TGATTCTTAGGTCCGAGAA..................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AATGAGTCCGGCCGAAGGCGC........................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................GACGGGTGCCGCTCATGCGTGT............................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCGTGTACTACCGGGCGAGGAG............................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TACGAGCTTCGGGCCAGACCGACG............................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................CGAGACACTGTGTGAGGGT........................................................ | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACACCCGTG.............................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................GCGTGTACTACCGGGCGAGG............................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCGTGTACTACCGGGCGAGGAGT............................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACAAA.................................................. | 20 | 2 | 9 | 0.78 | 7 | 6 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................ACTGTGTGAGGGAGACAC................................................... | 18 | 0 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACGCCC................................................. | 21 | 1 | 14 | 0.29 | 4 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..CTTCAGGACAAGCAGGGCACC....................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ACTGTGTGAGGGAGACAA................................................... | 18 | 1 | 8 | 0.25 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGAC..................................................... | 17 | 0 | 18 | 0.22 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACCGTGTGAGGGAGACACC.................................................. | 20 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ACTGTGTGAGGGAGACACC.................................................. | 19 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGATA.................................................... | 18 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................GGATGTCGCTCAAGCGTGT............................................................................................................. | 19 | 3 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CCCTGTGTGAGGGAGACAA................................................... | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGTCAA................................................... | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................ATCATGCGTGAAATACCGGG.................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................ATACACTGTGTGAGGGAGACGC................................................... | 22 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACGAAC................................................. | 21 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGACT.................................................... | 18 | 1 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGCGTGAGGGAGAC..................................................... | 17 | 1 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................CTACCGGGCGAGGAGTCGG......................................................................................... | 19 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................GACGGGTGCCGCTCATGC................................................................................................................. | 18 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACTGTGTGAGGGAGCCT.................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................ATGAACCCGGCTGAAGGCG......................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCGAAGTCCTGCGCGTCCCGTTGATGCTCGAAGCCCGGTCTGGCTGCCCACGGCGAGTACGCACATGATGGCCCGCTCCTCAGCCTACTAAGAATCCAGGCTCTGTGACACACTCCCTCTGTGGGCACTTTACTCAGGCCGGCTTCCGCGTATTTTCACAGGACCCGGCACCCA
*************************************(((((((((.(((((((((((.((..(((((.(((((((....)))))))..))))))))))).)))))))))...)))))))..........******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................AAGAATCCAGGCTCTGTGACACACT............................................................ | 25 | 0 | 1 | 6.00 | 6 | 5 | 1 | 0 | 0 |
| ..........................................................ACGCACATGATGGCCCGCTCCTCAGCCT........................................................................................ | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .CGAAGTCCTGCGCGTCCCGTTGAT..................................................................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................GGCCCGCTCCTCAGCCTACT..................................................................................... | 20 | 0 | 19 | 1.79 | 34 | 34 | 0 | 0 | 0 |
| .....................................................CGAGTACGCACATGATGGCCCGCTCCT.............................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................GGTCTGGCTGCCCACGGCGAGT.................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................CCACGGCGAGTACGCACATGGT......................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ............................................TGCCCACGGCGAGTACGCAGAT............................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................AATGATGGCCCGCTCCTCAGCCTACT..................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................GCTGCCCACGGCGAGTACGCACATGAT......................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................ACGGCGAGTACGCACATGATGGCC..................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................GCCCACGGCGAGTACGCACATGAT......................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................CACGGCGAGTACGCACATGAT......................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................CCGGTCTGGCTGCCCACGGCGACT.................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................GCCCACGGCGAGTACGCACATGA.......................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................CATGATGGCCCGCTCCTCAGCCTACT..................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..GAAGTCCTGCGCGTCCCGTTGAT..................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................GATGGCCCGCTCCTCAGCCTACT..................................................................................... | 23 | 0 | 19 | 0.68 | 13 | 12 | 1 | 0 | 0 |
| ...............................................................AAAGATGGCCCGCTCCTCAGCCT........................................................................................ | 23 | 2 | 15 | 0.40 | 6 | 6 | 0 | 0 | 0 |
| .................................................................TGATGGCCCGCTCCTCAGCCTACT..................................................................................... | 24 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................ATGGCCCGCTCCTCAGCCTACT..................................................................................... | 22 | 0 | 19 | 0.26 | 5 | 5 | 0 | 0 | 0 |
| ............................................TGCCCACGGCGAGTACGCACTTGAT......................................................................................................... | 25 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................GGCCCGCTCCTCAGCCTAC...................................................................................... | 19 | 0 | 19 | 0.21 | 4 | 4 | 0 | 0 | 0 |
| ................................................................AAGATGGCCCGCTCCTCAGCCTACT..................................................................................... | 25 | 1 | 15 | 0.20 | 3 | 0 | 3 | 0 | 0 |
| ........................................TGGCTGCCCAGAGCGAATA................................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................AAAGATGGCCCGCTCCTCAGCCTACT..................................................................................... | 26 | 2 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| ................................................................AAGATGGCCCGCTCCTCAGCCT........................................................................................ | 22 | 1 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................TCCAGGCTCGGTGACACACT............................................................ | 20 | 1 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................AACCAGGCTCGGTGACACACT............................................................ | 21 | 2 | 18 | 0.11 | 2 | 1 | 0 | 0 | 1 |
| ..................................................................GATGGCCCGCTCCTCAGCCTAC...................................................................................... | 22 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| .......................................................................CCCGCTCCTCAGCCTACT..................................................................................... | 18 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................ATGGCCCGCTCCTCAGCCTAC...................................................................................... | 21 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ..........................................GCTGCCCACGGCGAGTACGCAC.............................................................................................................. | 22 | 0 | 19 | 0.11 | 2 | 1 | 1 | 0 | 0 |
| ......................................................................GCCCGCTCCTCAGCCTACT..................................................................................... | 19 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................TGGCCCGCTCCTCAGCCT........................................................................................ | 18 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................TGGCCCGCTCCTCAGCCTACT..................................................................................... | 21 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ............................................................................................AAAACAGGCTCGGTGACACACT............................................................ | 22 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................AAAGATGGCCCGCTCCTCAGCCTAC...................................................................................... | 25 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................AAAGATGGCCCGCTCCTCAGC.......................................................................................... | 21 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ..................................................GGGCGAGTATGCACAAGATGG....................................................................................................... | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 |
| ...............................................CCACGGCGAGTACGCACAAGATG........................................................................................................ | 23 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ..................................................CGGCGAGTACGCACAAGAT......................................................................................................... | 19 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................AGAACCCAGGCTCGGTGACACACT............................................................ | 24 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................AGAGTCCAGGCTCGGTGACACACT............................................................ | 24 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................ATGGCCCGCTCCTCAGCCTA....................................................................................... | 20 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................GGCCCGCTCCTCAGCC......................................................................................... | 16 | 0 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................TTGGCCCGCTCCTCAGCCTAC...................................................................................... | 21 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................GATGGCCCGCTCCTCAGCC......................................................................................... | 19 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................CTGCCCACGGCGAGTACGCAC.............................................................................................................. | 21 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................AATGGCCCGCTCCTCAGCCTACT..................................................................................... | 23 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................TGCCCACGGCGAGTACGCA............................................................................................................... | 19 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................TGCCCGCTCCTCAGCCTACT..................................................................................... | 20 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................CTGCCCACGGCGAGTACGCACT............................................................................................................. | 22 | 1 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | Unknown_group_117:19626-19799 + | dps_118 | AGCTTCAGGACGCGCAGGGCAACTACGAGCTTCGGGCCAGACCGACGGGTGCCGCTCATGCGTGTACTACCGGGCGAGGAGTCGGAT---GATT-CTTAGGTCCGAGACACTGTGTGA---GGGAGACACCCGTGAAATGAGTCCGGCCGAAGG---CGCATAAAAGTGTCCTGGGCCGT----GGGT |
| droPer2 | scaffold_54:342155-342328 - | AGCCTCAGGACGCGCAGGGCAACTACGAGCTTCGGGCCAGACCGACGGGTGCCGCTCATGCGTGTACTACCGGGCGAGGAGTCGGAT---GATT-CTTAGGTCCGAGACACTGTGTGA---GGGAGACACCCGTGAAATGAGTCCGGCCGAAGG---CGCATAAAAGTGTCCTGGGCCGT----GGGT | |
| droAna3 | scaffold_9689:9619-9755 - | CGCG---------------------------------------------CGCTGCTC-----GGCACTACAACCTGAGGAGGCGGAAGTGGAGCCCGAAGATCGGCGACATGGTGTGGGCAAAGGAACACCATTTGTCTAAAG-CAGTCGAAGGTTTCGCGCCAAAGTACGACGGGCCGTACCAGTGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
Generated: 05/15/2015 at 02:36 PM