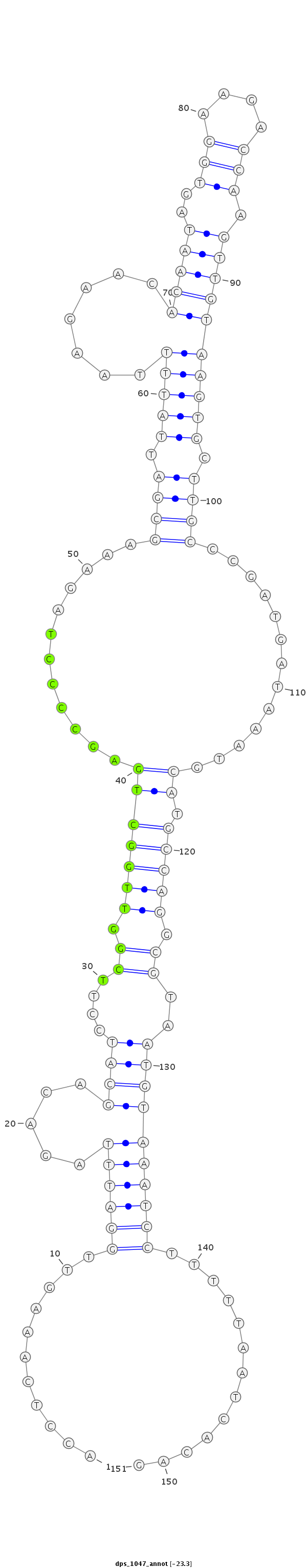
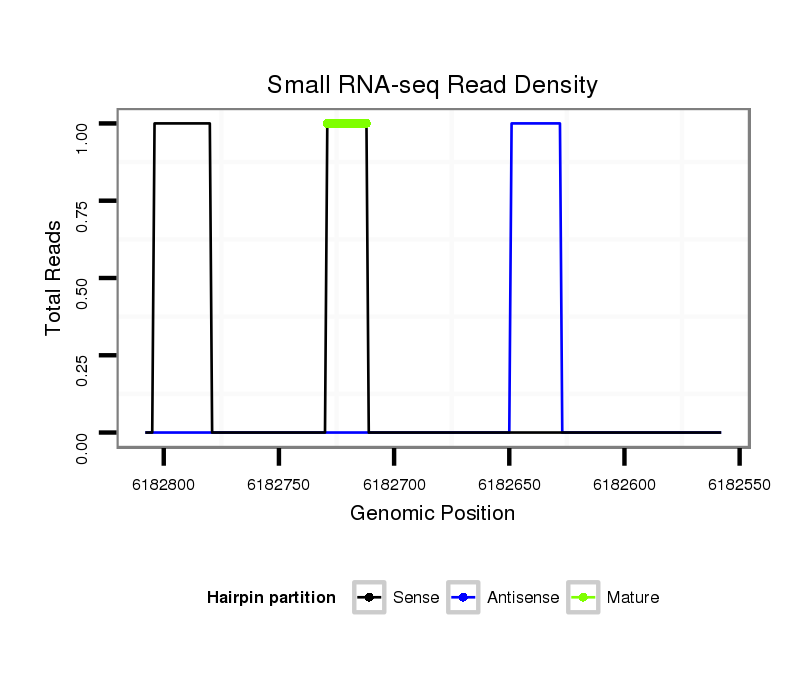
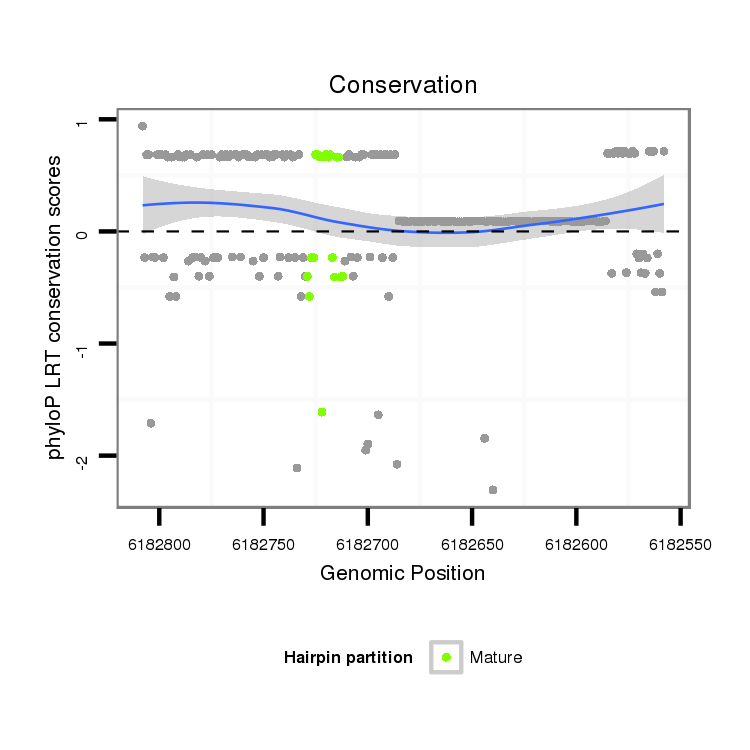
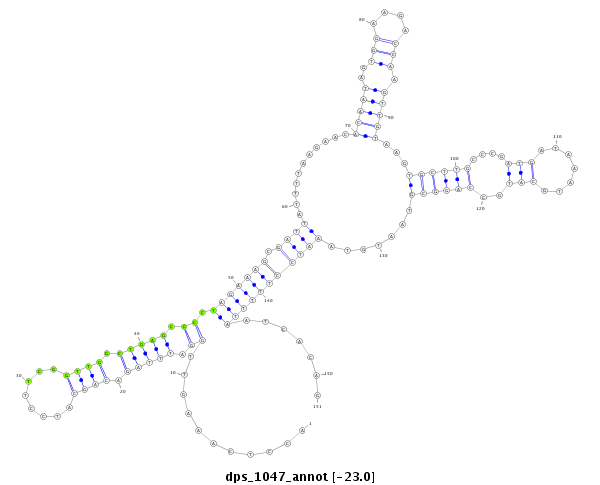
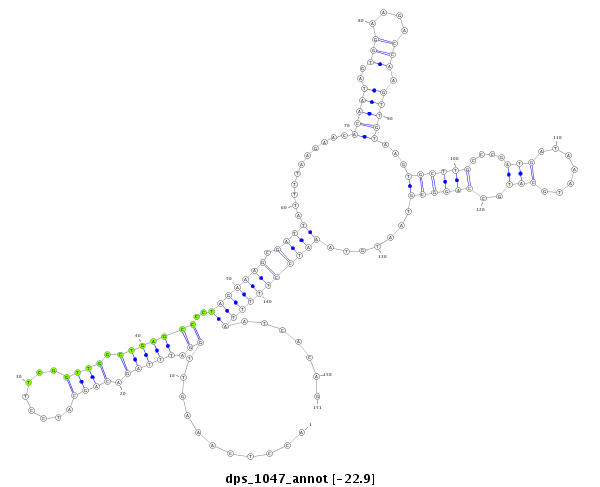
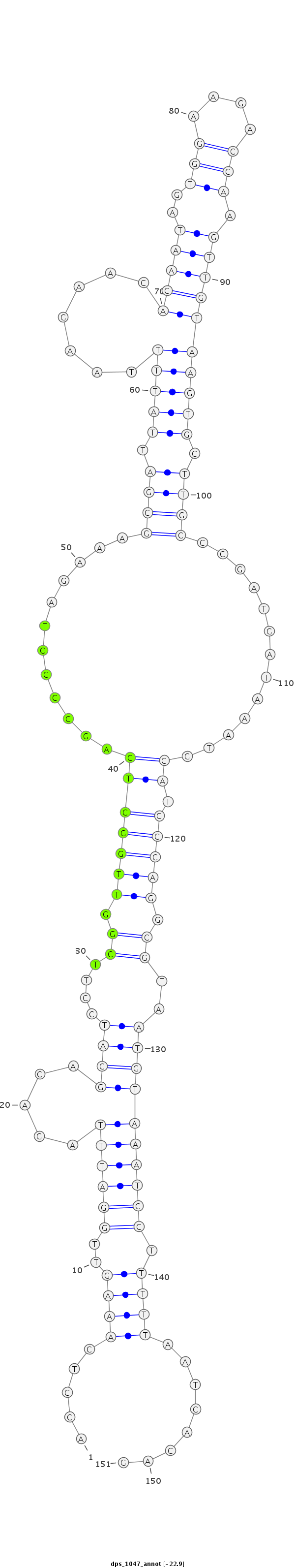
ID:dps_1047 |
Coordinate:3:6182608-6182758 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.0 | -22.9 | -22.9 |
 |
 |
 |
CDS [Dpse\GA21437-cds]; exon [dpse_GLEANR_14542:3]; intron [Dpse\GA21437-in]; Antisense to intron [Dpse\fat-spondin-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACTTTAGTTTGTCGGCGAGCGTTACAGAGTATATCCGGAGTGATTTGAAAACCTCAAAGTTGGATTTAGACAGCATCCTTCGGTTGGCTGAGCCCCTAGAAAGCGATTATTTTAAGAACACAATAGTGGAAGACCAAGTTGTAAGTGCTTGCCCGATGATAAATGCATGCCAGGCGTAATGTAAATCCTTTTTAATCACAGAAAAACGCCACAAAAATGGGAAGCCCACAGAAGGAGTTTACTAAACTCCA **************************************************...........((((((.....((((....((.(((((((............((((.(((((.......(((((..(((....))).)))))))))).)))).............)).))))).))..)))))))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
GSM343916 embryo |
GSM444067 head |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................TTGGACCGCCTCCTTCGGTTGGC................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....TAGTTTGTCGGCGAGCGTTACAGAG.............................................................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................TCGGTTGGCTGAGCCCCT.......................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGAAGACGAACTTGTAAGTCC....................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................ATACGCAGAGTGATTTGAAAA........................................................................................................................................................................................................ | 21 | 3 | 6 | 0.33 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..............................ATACGCAGAGTGATTTGAAA......................................................................................................................................................................................................... | 20 | 3 | 10 | 0.30 | 3 | 2 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................ACGTTGTAAGTGGTTGACC................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TGAAATCAAACAGCCGCTCGCAATGTCTCATATAGGCCTCACTAAACTTTTGGAGTTTCAACCTAAATCTGTCGTAGGAAGCCAACCGACTCGGGGATCTTTCGCTAATAAAATTCTTGTGTTATCACCTTCTGGTTCAACATTCACGAACGGGCTACTATTTACGTACGGTCCGCATTACATTTAGGAAAAATTAGTGTCTTTTTGCGGTGTTTTTACCCTTCGGGTGTCTTCCTCAAATGATTTGAGGT
**************************************************...........((((((.....((((....((.(((((((............((((.(((((.......(((((..(((....))).)))))))))).)))).............)).))))).))..)))))))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................ATTTACGTACGGTCCGCATTAC...................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..................................................................................................................................................CGGACGAGCTACTTTTTACGT.................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 |
| .................................................................................................................................................................................TTACATTTAGGAAATATTC....................................................... | 19 | 2 | 5 | 0.20 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 3:6182558-6182808 - | dps_1047 | ACTTTAGTTTGTCGGCGAGCGTTACAGAGTATA------------------------------------------------------TCCGGAGTGATTTGAAAACCTCAAAGTTGGATTTAGACAGCATCCTTCGGTTGGCTGAGCCCCTAGAA---------AGCGATTATTTTAAGAACACAATAGTGGAAGACCAAGTTGTAAGTGCTTGCCCGATGATAAATGCATGCCAGGCGTAATGTAAATCCTTTTTAATCACAGAAAAACGCCACAAAAATGGGAAGCCCACAGAAGGAGTTTACTAAACTCCA |
| droPer2 | scaffold_2:6397421-6397671 - | ACTTCAGTTTGTCGGCGAGCGTTACAGAGTATA------------------------------------------------------TCCGGAGTGATTTGAAAACCTCAAAGTTGGATTTAGACAGCCTCCTTCGGTTGACTGAGCCCCTAGAA---------AGCGATGCTTTTGAGAACACACTAGTGGAAGACCAAGTTGTAAGTGCTTGCCCGATGATAAATACATCCCAGGCGTAATGTAAATCCTTTTTAATCACAGAAAAACGCCACAAAAATGGGAAGCCCACAGAAGGAGTTTACTAAACTCCA | |
| droMoj3 | scaffold_6496:23954816-23954844 - | A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCACACAGACGGAGCAGGAAAAAGCAGA | |
| droGri2 | scaffold_15112:987479-987663 + | ATTTTGATTTATCCGACAGCGTAATGGCATTTCTACAGGTAAGTCATATCTTTTGCACCTCTATTGAGCAGATTAATCTGTTTATTTTTTAGAGTGACTTGGAAACCACACAATTGGATGCAGATAGTTTGTGGGAATTGGCTGAAACCAGTGAGAAGCGCAACCGTGATTCCTTTGAAAAGATA--------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 11:52 PM