| dp5 |
3:5960640-5960890 + |
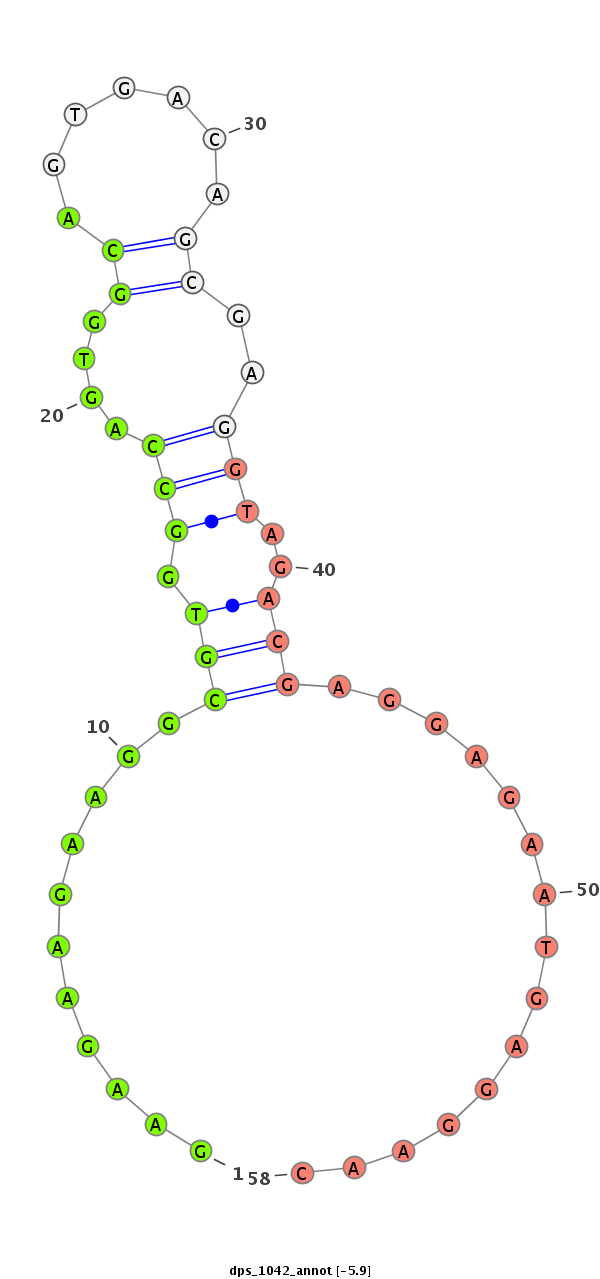
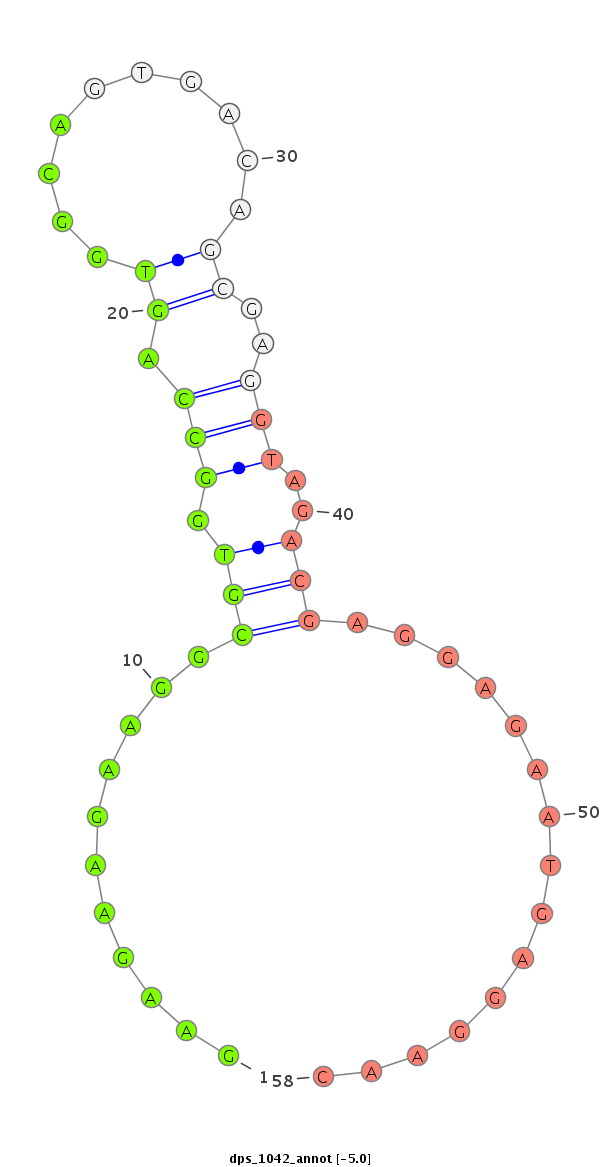
dps_1042 |
ATCAGGC------------------------GTCATCTCCAGTGCGTAC---AGCGACAGCAGCGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTG---GCAGTGACAGCGAGGTAGACGAGGAGAATG---------AGGAACAGGGGGC------AGAAGAGGA---AGA---------------TC------A------------------------------------------------------GCAGCAGCA----GGCAG------------AGGTAGAGG---------------AA----------------------------------------------------------------------------------------------------------------------GTCGAAAACGACTCCGAAGA---------TTCATCG------GAATATGAAGAAGTAACAGTCACCGAAAGCAGCGA---------------TGAAG---AAGAG |
| droPer2 |
scaffold_2:6161596-6161846 + |
dpe_1086 |
ATCAGGC------------------------GTCATCTCCAGTGCGTAC---AGCGACAGCAGCGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTG---GCAGTGACAGCGAGGTAGACGAGGAGAATG---------AGGAACAGGGGGC------AGAAGAGGA---AGA---------------TC------A------------------------------------------------------GCAGCAGCA----GGCAG------------AAGTAGAGG---------------AA----------------------------------------------------------------------------------------------------------------------GTCGAAAACGACTCCGAAGA---------TTCATCG------GAATATGAAGAAGTAACAGTCACCGAAAGCAGCGA---------------TGAAG---AAGAG |
| droWil2 |
scf2_1100000004510:2489658-2489926 + |
dwi_441 |
ACCAGGC------------------------AGCATCTCCAGTGCGTAC---AGCCACGACAAGCGCGCGAGCCGAGGTAAGTGTGAATGAGGCAAAT---GAA---------------------------------GAAGAAGATCTGTCCAGTGA---CAGTGGCAGCGAGGAAGAGGACGACGAAG---------AGGAAG---------------------------------------------------------------------ATGATGCAG---------CAGTTGAACAG---------------AA----------------------------AGAAGTAGTGCCACAGGTACA----------------------------------------------------------------------------------------------------AGAGGAGGAA------AAGGAAAATGAGGATGAAGCTTCAACATCCTCATCAGAGTATGAATACGAAGAGGTTACAGCCACTGAGAGTGAGTC---------------AGAAG---AAGAG |
| droVir3 |
scaffold_12875:19589744-19590036 - |
dvi_7992 |
ATCAGTCGGCGACA------------GCGGCGGCATCTCCAGTGCGTACAACGACAGCGTCGACAGCGCGAGCCGAGGTAAGTGTGAATGAAACAAAT---GAA---------------------------------GAAGAAGGCGTGGCCAGTGACAACAGTGGCAGCAGCGAGACGGAGAGTGAGACGGGAAGTGA---GAGCGAGGC------GGAGGAAGA---AGA------GCAGCAGCAGC------A------------------------------------------------------GCAGCAGCA----GGCGC---------AGTTGGCGCAGG---------------CACC----------------------------------------------------------------------------------------CGCTGAGGATCAAGTGGATAACG------------------------A------TTCCAGCTCCGAGTATGAATACGAGGAGGTGACAGCCAGTGAGAGTGAGC------------------------AGAGT |
| droMoj3 |
scaffold_6496:6640169-6640509 - |
|
ATCAGGCAGCGACAGCAGCAGCGGCAGCGGCGGCATCTCCAGTGCGTAC---AGCAACGGCGACAGCGCGAGCCGAGGTAAGTGTGAATGAGACAAATGAAGAAGATGAGGAAGCAGATGAGGAAGAAGAAGCAGAAGCAGAAGAGGAAGCCAGTGACAGCAGTGACAGCAGCGAAGCTGAGACTGGGACCGAAAGTGA---GAGTGAGAG---T---GAGAGTGAGACAGA------GCAACAGCAGC------A------------------------------------------------------GCAGCAGCA----GGCAG---------AACAA---------------------------------------------------------CA-------CCAGCTGCAGCTGCCAGAGCA---------------------------------------GGTGGATAATG------------------------C------GACCAGCTCT------GAGTATGAAGAGGTGACAGTCAGTGAGAGTGAGGA---------------G---AGCGACGAG |
| droGri2 |
scaffold_15245:17961623-17961948 + |
|
ATCAAGC---GACA------------ACAGCATCATCTTCAGTGCGTACAGCAACAACGGCGACAGCACGAGCCGAGGTAAGTGTGAATGAAACAAATGAAGAAGAAGAAGTGGCAGCAGCAGCAGAAAATGAT---GAAGTAGGTGTGGCCAGTGACGACAGTGGCAGCAGCGACATTGAGAGCGAAA---------CAGAAC---A------------------------------------------------------------------------AGAGCGTGAACAAGTTCAAGAGCAAGAGTTGCAACCACA----AGCAG------------TAG---------------------------------CGGTGATAAAT-------------------------------------------------------------------------GAAGATCAAGTGGACAACGACGACGTCGATGACG---------C------CTCTTCCTCTGAATATGAGTATGAAGAGGTGACAGCGAGTGAGAGCGAAG------------------------AGGAG |
| droAna3 |
scaffold_13266:17449042-17449349 - |
dan_2079 |
ATCAGGC------------------------GGCGTCTCCAGTGCGTAACAGTGCAGCAACCGCGACGCGGGCCGAGGTAAGTGTGAATGAGACAAATACAGAA---------------------------------GAAGAAGCCCTGGCCAGCGAAGGCAGTGATAGCGAAGAGGAGGAGGAAGAAA---------CAGAAG---------------------------------------------------------------------AGTCTGAAGAAAGTGAAGTAGA---ACAG---------------GA-------------------------------------------------GGCAG------------GTGTGGCGCCGCAGGA-------CACGCTTAATGTTCCAGAA---GCTAGCCA---GGATGCCTCATCGGGTTCCGAGGTTACAGAAGAGGAGG------------------------A------ATCCTCTTCC------GAATACGAAGAAGAAACAGCAGAAGAAAGCGAAGA---------------A---GAGGAAGAA |
| droBip1 |
scf7180000396735:710634-710938 - |
|
ATCAGGC------------------------GGCGTCTCCAGTGCGTAACAGTGCAGCAACCGCGACGCGGGCCGAGGTAAGTGTGAATGAGACAAATACAGAA---------------------------------GAAGAAGCCATAGCCAGCGAAAGCAGTGACAGCGAAGAGGAGGAGGAAGAAA---------CAGAAG---------------------------------------------------------------------AGTCTGAAGAAAGTGAAGTAGA---ACAG---------------GA-------------------------------------------------GGCAG------------AAGTAAGGCCACAGGA-------CCAGCTTGATGTGCCAGAA---GCTAGCCA---GGATGCCTCGTCGGCATCCGAGGTAACAGAGGAGGAAG------------------------A------ATCCTCTTCT------GAATACGAAGAAGAAACAGCAACCGAAAGCGAAG------------------AAG---AAGGG |
| droKik1 |
scf7180000302682:642206-642516 + |
|
CTCAGGC------------------------GGCGTCTCCAGTGCGTAATTCGGCGGCAGCGGCAACGCGAGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGACAGCAGTGACAGCGAGGAAGAGGAGGAGGATG---------ATGAGACTGAGGA------GG------A---AGA---------------T-------------------------------------------------------------------------------G---------ATGAAGTAGAAG---------------AGTCTCCTG------------AGGTC-CTGCCAGAGTTGGCCCAAGGTCTTAGCCCCCCAGAAGTGCCT---GCAGAACCCACATC------CTCTGAACAAACCGAGGAAGAAG------------------------A------GTCTTCCTCG------GAGTATGAAGAAGTAACAGCCACTGAAAGCGAAG------------------AGG---AGGAG |
| droFic1 |
scf7180000454044:681139-681515 + |
|
ATCAGGC------------------------GGCGTCTCCAGTGCGTAACAGTGCCGCATCGACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAATGACAGCAGTGACAGCGAAGAAGAAGTTGAAGAGG---------AAGGGGAGGAGGAGGAG---GAGG---A------GGAGGAGGAGGAGGAGG------AGGAGGAGGAGGAAGAAGAATCAGCAGGGGAAGG----------------------------------GGTAG---------AACAGGTGGAGG---------------AACCTTCAGTGGGTGTGGTAGAAGAGGCTTCAAGAGA-------AGTTCTTAATCTACCAGAAAAGGCT---TCGAAAGCCACTTC------TTCGAAAGTTCCCGAAGAAGAAG------------------------A------ATACTCTTCC------GAATACGAGGAAGTAACGGCCAACGAAGAGAGTGA---------------AGAAG---AAGAA |
| droEle1 |
scf7180000491228:23762-24078 - |
|
ACCAGGC------------------------GGCGTCTCCAGTGCGTAATAGTGCGGCATCTGCGACGCGGGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGACTGCAGTGACGGCGAAGAGGAAGAAGAAGAGG---------AAGAGG----------------------------------------------------------------------------------------------------------------------------------------AGGGGGAAG---------------AATCTGTAGAGGCGGAAATCGAAGAGGCTCCCAGGGA-------ATCTCTTAACCTTCCAGAGGAATCT---CTGCAAGCAACCTC------TTCCGAAGAAACCGAAGAGGAAG------------------------A------ATCCTCATCG------GAATACGAAGAAGAAACAGCCACCGAAAGTGAAGAGGATGGGGAGGGGGGAGAGG---AAGAG |
| droRho1 |
scf7180000779618:14865-15163 + |
|
ACCAGGC------------------------AGCGTCTCCAGTGCGTAATAGTGCGGCGACTACGACGCGAGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGACAGCAGTGACAGCGAAGAAGAGGGGGAGGAGG---------AGGAGG---------------------------------------------------------------------AGGAT-------------------------------------------------G---------AGGAGGGGGAGG---------------AATCCGTAC------------AGGAAGCTACCGGAAA-------TGTGCTTAACCTTCCAGAAGAAGCT---CTTCAAGCTACATC------TTCCGAAGAAACGGAGGAGGAAG------------------------A------GTCCTCTTCC------GAATACGAAGAAGTCACAGCCACCGAGAGTGAAGA---------------A---GAAGAAGAG |
| droBia1 |
scf7180000302292:1478816-1479105 - |
|
ACCAGGC------------------------GGCGTCTCCAGTGCGTAACAGTGCAGCCTCTACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAGGAAGAGGAGGAGGAGG---------AGG----------------------------------------------------------------------------------------------------------------------CGGGGGGAG---------AGCAACCGGAAG---------------AATCTGTTC------------AAGTGGTTCTACAGGA-------TGTGCTTAGTCTA---GGAGAGCCT---CAGCAGCCCACGTC------TTCCGAAGAAACGGAGGAGGAAG------------------------A------GTCCTCCTCC------GAGTACGAGGAAGTAACAGCTAGCGAAACTGAAG------------------------AGGAA |
| droTak1 |
scf7180000415991:355332-355651 + |
|
ATCAGGC------------------------GGCGTCTCCAGTGCGTAACAGTGCGGCCTCAACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAGAGCAGTGACAGCGAAGAAGAGGAGGAGGAGG---------AGGATG---AGGA------GGAAGAATC---GGT---------------AG------A---G------------------------------------------------------G----AGGGGGCAG---------AACAAGCTGAAG---------------AATCTGTAG------------AAGGGGCTCCTAGGGA-------ATTGCTTAATCCCCCAGAAGAATCT---CTGCAGCCCACGTC------TTCCGAAGTAACAGAGGAGGAAG------------------------A------ATCGTCCTCC------GAGTATGAAGAAGTAACAGCCACCGAAAGTGAGG------------------AAG---AACAG |
| droEug1 |
scf7180000409663:443229-443533 - |
|
ACCAGGC------------------------AGCGTCTCCAGTGCGTAATAGTGCCGCAGCTACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGAAG---------AAGAAG---------------------------------------------------------------------AGGAT------------------------------------G----AGGGTCCAG---------AACAAGCAGAAG---------------AATTTGCAG------------AAGTGGCTTCTAGGGA-------CCGGCTTAACCCACCGGAGGAATCC---CTGCAACCGACCTC------TTCCGAAGTAACGGAGGAGGAGG------------------------A------GTCCTCTTCT------GAGTACGAAGAAGTCACAGCCACCGAAAGTGAAG------------------AAG---AGCAG |
| dm3 |
chr2R:14885876-14886201 + |
|
ACCAGGC------------------------GGCGTCCCCAGTGCGTAATAGTGCGGCATCCACGACGCGAGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGATGAGGAAGTTG---------AAGATG---AAGATGAAGAAGAAGAGGT---AGA---------------CG------A----------------------------------------------------------------GGAGGAGG---------AACAAGTAGAAG---------------AATCTGTAGAAG------AAGAAGAGGCTCCCAGGGA-------ATTGCTGAACCTACCAGAGGAAGCT---CTGCAACCGACCTC------ATCTGAAGTTACAGAGGAGGAAG------------------------A------ATCCTCTTCC------GAATACGAAGAAGTAACGGCCACTGAAAGTGAAG------------------------AAGAG |
| droSim2 |
2r:15534852-15535168 + |
dsi_25601 |
ACCAGGC------------------------GGCGTCCCCAGTGCGTAATAGTGCGGCATCCACGACGCGAGCAGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGAGG---------AAGAAG---AAGA------AGATGAGGT---GGA---------------GG------A---G------------------------------------------------------G----AGGAGGAAG---------AACAAGTAGAGG---------------AATCTGTAG------------AAGGGACTCCTAGGGA-------ATTGCTTAACCTACCAGAGGAAGCT---CTGCAACCGACCTC------GTCTGAAGTTACAGAGGAGGAAG------------------------A------ATCCTCTTCC------GAATACGAAGAAGTAACAGCCACTGAAAGTGAAG------------------------AAGAG |
| droSec2 |
scaffold_1:12392163-12392482 + |
|
ACCAGGC------------------------GGCGTCCCCAGTGCGTAATAGTGCGACATCCACGACGCGAGCAGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAAGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGAGG---------AAGAAG---AAGA------AGAAGAGGT---GGA---------------GG------AGGAG------------------------------------------------------G----AGGAGGAAG---------AACAAGTAGAGG---------------AATCTGTAG------------AAGGGACTCCTAGGGA-------ATTGCTTAACCTACCAGAGGAAGCT---CTGCAGCCGACCTC------TTCTGAAGTTACAGAGGAGGAAG------------------------A------ATCCTCTTCC------GAATACGAAGAAGTAACAGCCACTGAAAGTGAAG------------------------AAGAG |
| droYak3 |
2R:14307770-14308113 - |
dya_571 |
ACCAGGC------------------------GGCGTCTCCAGTGCGTAGTAGTGCGGCATCCACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGATGAAGAGGAGGAGGATG---------TAGAGGAGGAAGATGAAGAAGAAG---------ACGAGGAGGAGGA---AGATGATGAGGAG------------------------------------------------------G----AGGGTGAACAACAAGTAGAACAA---------------------------GTAG------------AAGAAACGGGTAGGGA-------ATTGCTTAACCTCCCGGAAGAGTCA---CTGCAACCGACCTC------TTCTGAAGCAACAGAGGAGGAAG------------------------A------ATCCTCTTCC------GAGTACGAAGAGGTAACAGCCACTGAGAGTGAAGA---------------C---GAGCAGGAG |
| droEre2 |
scaffold_4845:9081439-9081782 + |
der_1081 |
ACCAGGC------------------------GGCGTCTCCAGTGCGTAATAGTGCGGCAACTACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAGGAAGAGGATGAGGAGG---------TGGAGGAGGAGGAGGAA---GAAG---A------CGAAGAGGAAGA---CGAGGATGAGGAG------------------------------------------------------G----AGGGGGAAG---------AACAAGCGGAGG---------------AATCTGTAG------------AAGATACGGGTAGGGA-------ACTGCTTAACCTCCCAGAAGAAGCT---CTGCAGCCGACCTC------TTCTGAAGTCACAGAGGAGGAAG------------------------A------ATCCTCATCG------GAGTACGAAGAGGTGACAGCCACTGAGAGTGAAGA---------------A---GAGCAGGAG |