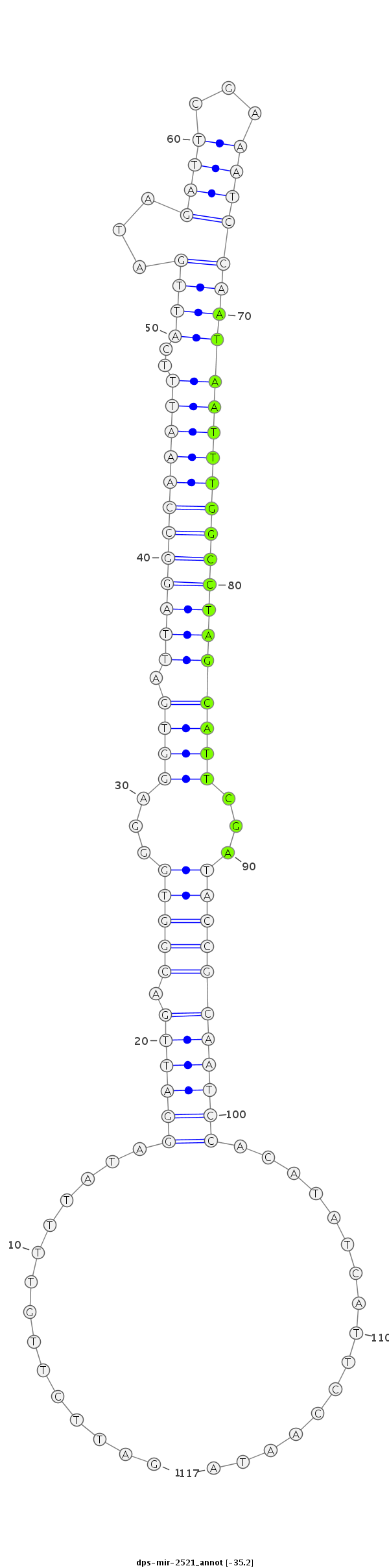
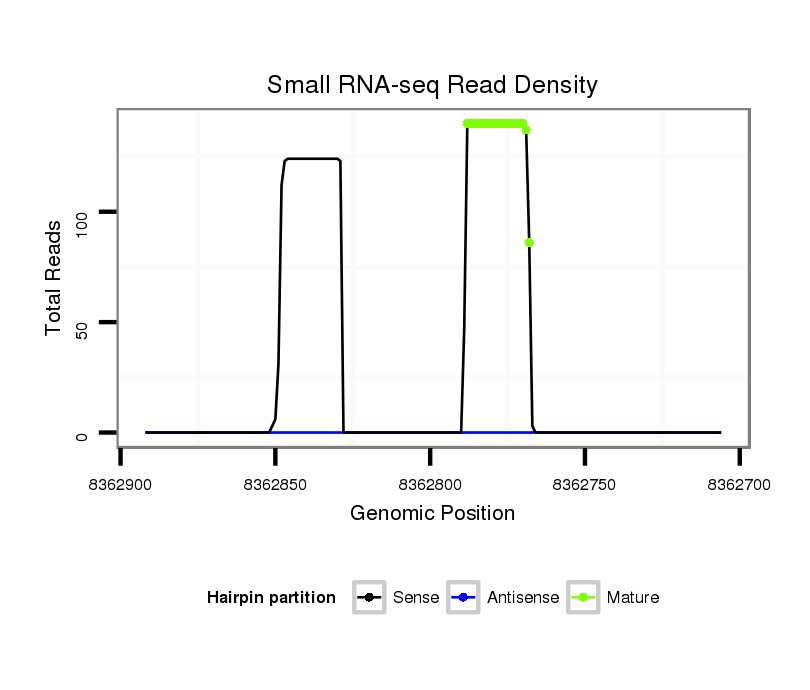
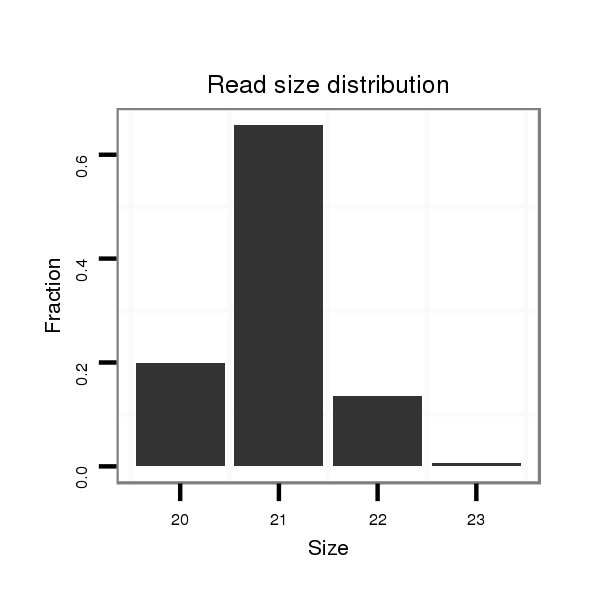
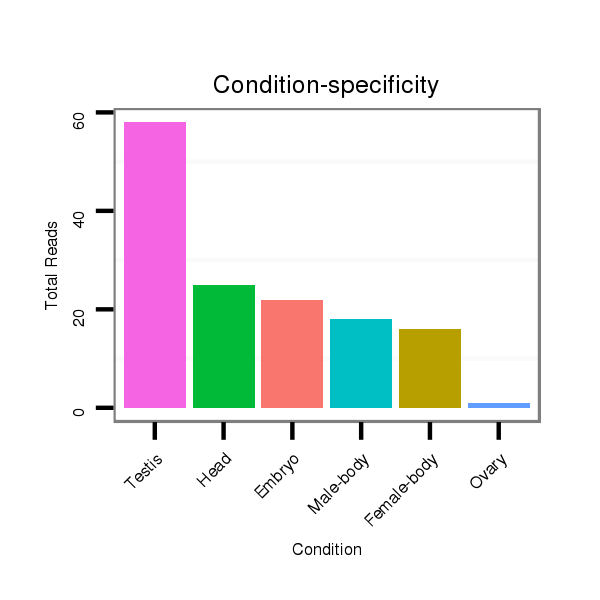
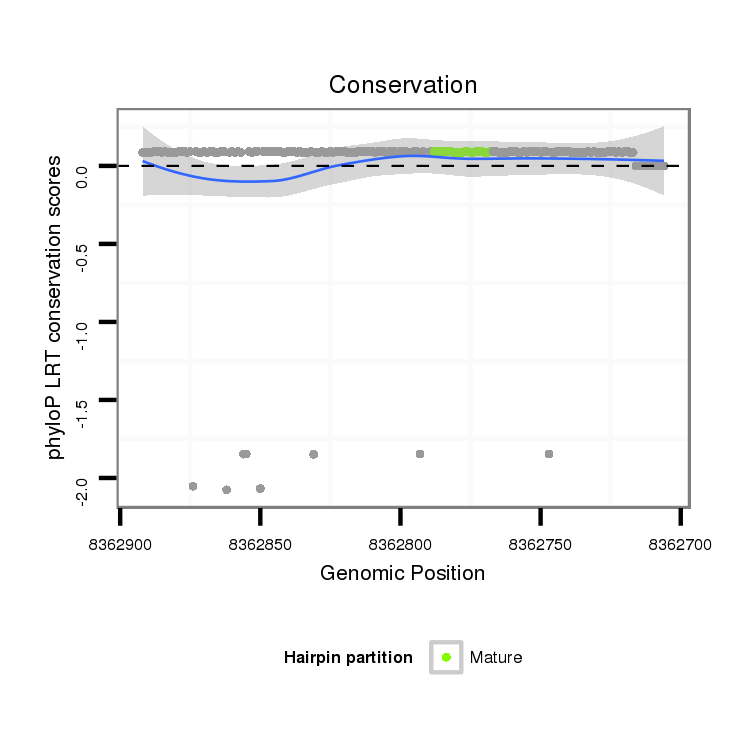
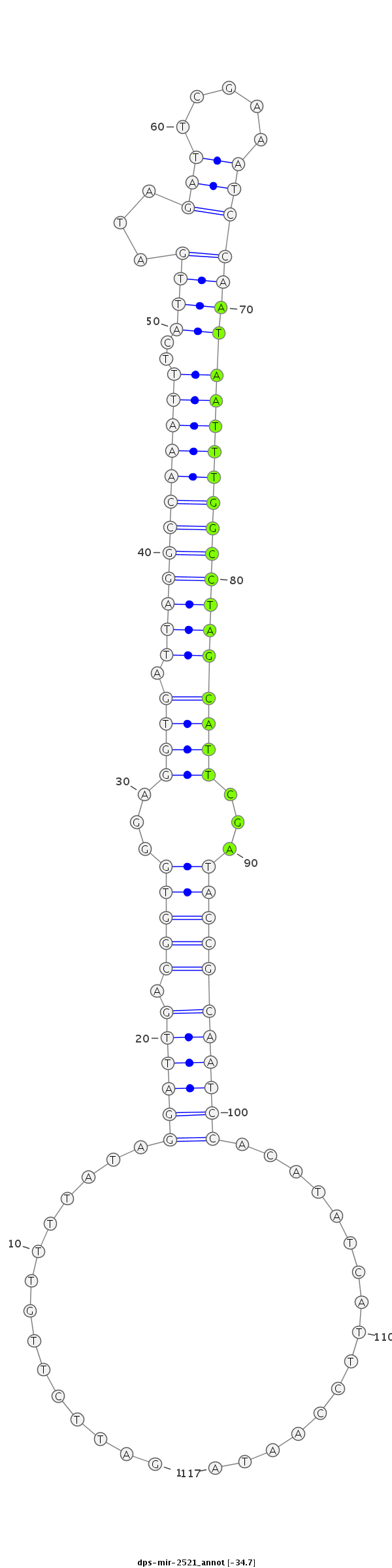
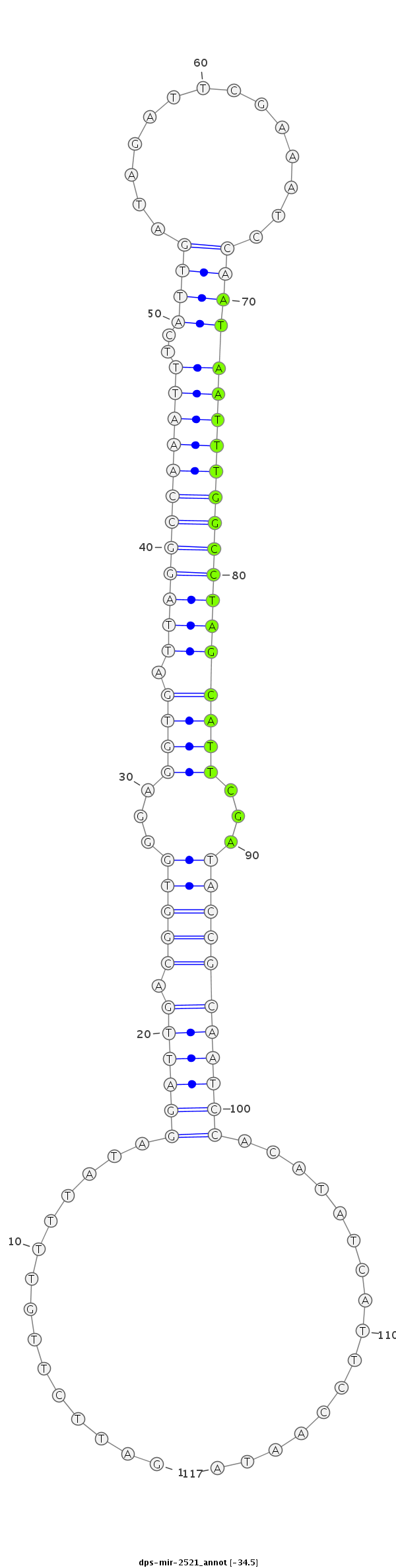
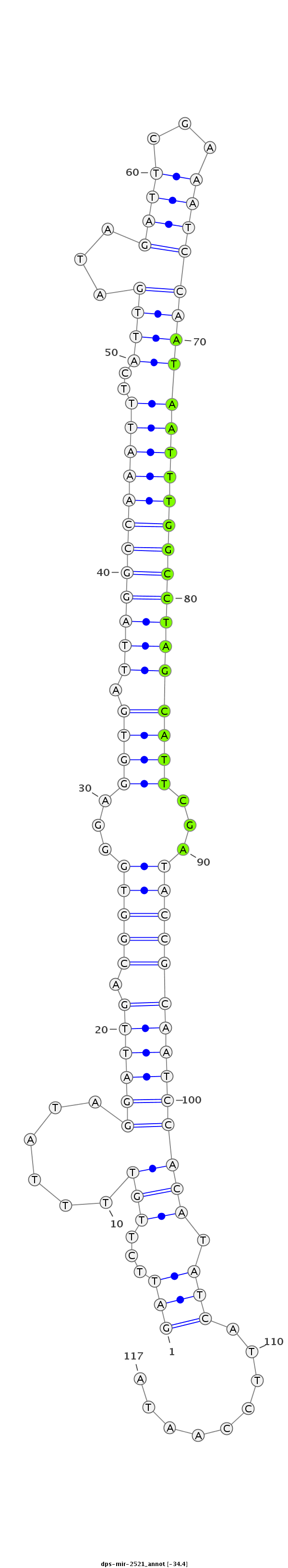
ID:dps-mir-2521 |
Coordinate:XL_group1e:8362756-8362842 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.7 | -34.5 | -34.4 |
 |
 |
 |
intergenic
No Repeatable elements found
|
GCCGTTTCACCCCAATCTCTCGTGTACGATAGTGAGATTCTTGTTTTATAGGATTGACGGTGGGAGGTGATTAGGCCAAATTTCATTGATAGATTCGAAATCCAATAATTTGGCCTAGCATTCGATACCGCAATCCACATATCATTCCAATATCCCGCGAGTGCCCATACTGTTGGGAGTTGATTTC
***********************************...............((((((.(((((...((((.((((((((((((..((((...((((...))))))))))))))))))))))))...)))))))))))................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
V112 male body |
GSM444067 head |
GSM343916 embryo |
M040 female body |
M062 head |
M059 embryo |
SRR902010 ovaries |
SRR902012 CNS imaginal disc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................TTTATAGGATTGACGGTGGG........................................................................................................................... | 20 | 0 | 1 | 80.00 | 80 | 78 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................ATAATTTGGCCTAGCATTCGA.............................................................. | 21 | 0 | 1 | 66.00 | 66 | 36 | 5 | 5 | 9 | 3 | 1 | 6 | 1 | 0 |
| .......................................................................................................AATAATTTGGCCTAGCATTCG............................................................... | 21 | 0 | 1 | 26.00 | 26 | 9 | 3 | 3 | 3 | 5 | 3 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCCTAGCATTCG............................................................... | 20 | 0 | 1 | 25.00 | 25 | 7 | 6 | 4 | 0 | 4 | 4 | 0 | 0 | 0 |
| ...........................................TTTTATAGGATTGACGGTGGG........................................................................................................................... | 21 | 0 | 1 | 25.00 | 25 | 24 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATAATTTGGCCTAGCATTCGA.............................................................. | 22 | 0 | 1 | 17.00 | 17 | 6 | 1 | 2 | 3 | 4 | 1 | 0 | 0 | 0 |
| .............................................TTATAGGATTGACGGTGGG........................................................................................................................... | 19 | 0 | 1 | 11.00 | 11 | 10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCCTAGCATTCGAA............................................................. | 22 | 1 | 1 | 5.00 | 5 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................TGTTTTATAGGATTGACGGTGGG........................................................................................................................... | 23 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................GTTTTATAGGATTGACGGTGGG........................................................................................................................... | 22 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATAATTTGGCCTAGCATTC................................................................ | 20 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................TAATTTGGCCTAGCATTCGAAA............................................................ | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCCTAGCATTCGAT............................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................CTCTAGTGACCGATAGTGA........................................................................................................................................................ | 19 | 3 | 13 | 1.38 | 18 | 0 | 1 | 0 | 0 | 17 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGCCCTAGCATTCGA.............................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TTTTATAGGATTGACGGGGGG........................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCGTAGCATTCGA.............................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATAATTTAGCCTAGCATTCG............................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATAATTTGGCCTAGCATTCGAT............................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................ATTAATTTGGCCTAGCATTCGA.............................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................AGATTTTGGCCTAGCATTCGAT............................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................ATATTCTGGCCTAGCATTCGA.............................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TATAGGATTGACGGTGGG........................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ACAATTTGGCCTAGCATTCGA.............................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................AATAAGTTGGCCTAGCATTCG............................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................AATCATTTGGAGTAGCATTCGA.............................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTTGGCCTAGCATTCGA.............................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................TATAGGATTGACGGGGGG........................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCCGAGCATTCG............................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCCTAGCAGTTGA.............................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTTATAGGATTGACGGTGG............................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATAATTTGGCCTAGCATTCGAGA............................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TTTTATAGGCTTGCCGGTGGG........................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................TGAGATTCTTCTTTTATAGTTT..................................................................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGAATTTGGTCTAGCATTCGT.............................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............TCTCTAGTGACCGATAGTG......................................................................................................................................................... | 19 | 3 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATTCGATACAGCACCCCACA................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............TCTCTAGTGACCGATAGTGA........................................................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GCCGTGTCTCCCCTATCTCT....................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TCTAGTGACCGATAGTGA........................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CGGCAAAGTGGGGTTAGAGAGCACATGCTATCACTCTAAGAACAAAATATCCTAACTGCCACCCTCCACTAATCCGGTTTAAAGTAACTATCTAAGCTTTAGGTTATTAAACCGGATCGTAAGCTATGGCGTTAGGTGTATAGTAAGGTTATAGGGCGCTCACGGGTATGACAACCCTCAACTAAAG
***********************************...............((((((.(((((...((((.((((((((((((..((((...((((...))))))))))))))))))))))))...)))))))))))................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
GSM444067 head |
M062 head |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TGGCGATAGGTGTGGAGTA.......................................... | 19 | 3 | 20 | 17.15 | 343 | 211 | 115 | 14 | 3 | 0 |
| ..............................................................................................................................TGGCGATAGGTGTAGAGTAGGGT...................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGATAGGTGTGTAGTAGGG....................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGTTAGGTGTGGAGT........................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................TGGCGATAGGTGTGGAGTAA......................................... | 20 | 3 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................CGTTAGGTCGATAGTATGGTT..................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGATAGGTGTGCAGTA.......................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGATAGGTGTGGAGTAAG........................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGTTAGGTGTGGAGTAG......................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................TAGGTGTACAGTAGGGTTGT................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................TGGCGCTAGGTGTGGAGTA.......................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGATAGGTGTGTAGTAG......................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TTTAGGTTCTTAAACTGG........................................................................ | 18 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................CCGGTTGATGGTAACTATC............................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGCGATGGGTGTAGAGTA.......................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| dp5 | XL_group1e:8362706-8362892 - | dps-mir-2521 | Known_mirbase | GCCGTTTCACCCCAATCTCTCGTGTACGATAGTGAGATTCTTGTTTTATAGGATTGACGGTGGGAGGTGATTAGGCCAAATTTCATTGATAGATTCGAAATCCAATAATTTGGCCTAGCATTCGATACCGCAATCCACATATCATTCCAATATCCCGCGAGTGCCCATACTGTTGGGAGTTGATTTC |
| droPer2 | scaffold_13:201553-201728 + | dpe_336 | confident | GCCGTTTCACCCCAATCTATCGTGTACGATCGTGAGGCTCTTTTTTTATAGGATTGACGGTAGGAGGTGATTAGGCCAAATTTCATTGATAGATTCGAAGTCCAATAATTTGGCCTAGCATTCGATACCGCAATCCACATATCATCCCAATATCCCGCGAGTGCCCATACTGTTGG----------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 10/20/2015 at 07:04 PM