

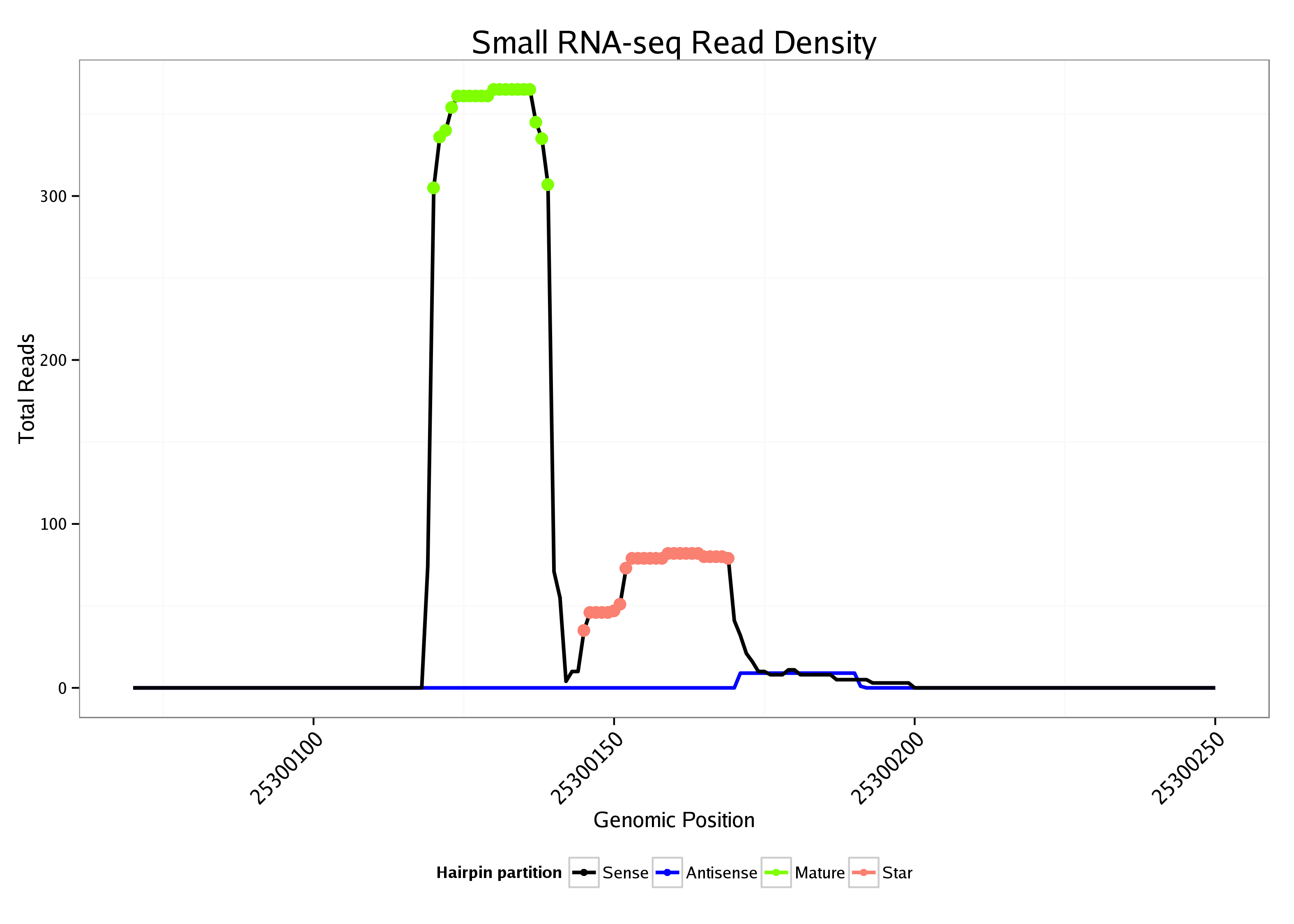
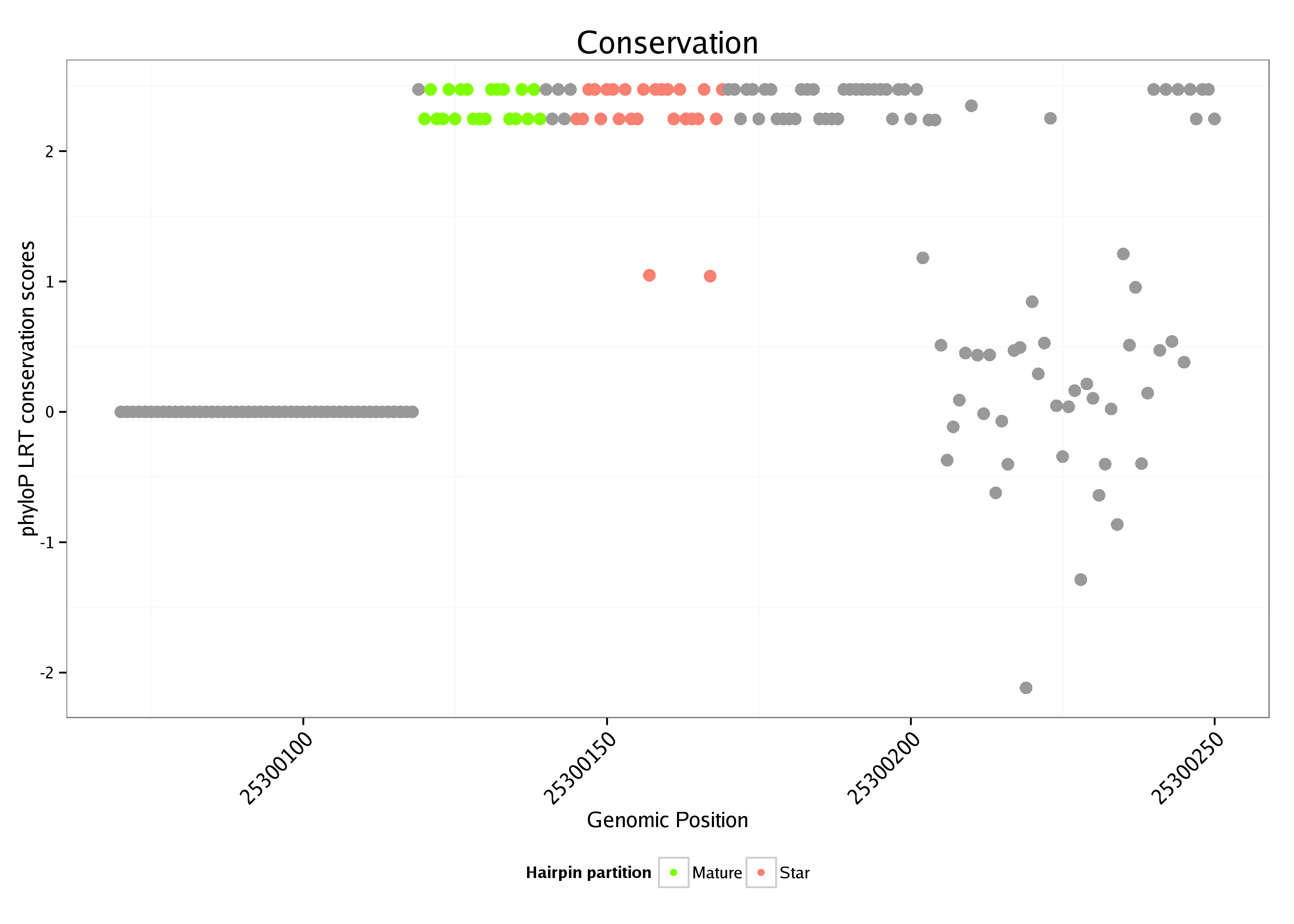
ID:dme_6 |
Coordinate:chrUextra:25300120-25300200 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -13.2 | -12.3 | -12.2 |
 |
 |
 |
unknown
|
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGTGGAGAAGGGTTTCGTGTGACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCGAAAATTTTCAAGTAAAACAAAAATGCCTAACTATATAAACAAAGCGAATTATAATACACTTG
***********************************................((((...(((..(((..((((.(((......)))..))))..))).)))...))))........****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V085 CME W2 wing disc |
V135 CME W2 (wing disc line) |
V026 1182-4H cell |
V147 1182-4H cell |
V009 CMEL1 |
V126 CME L1 |
V028 CMEW1 Cl.8+ cell |
V033 CMEW1 Cl.8+ cell |
V091 fGS/OSS total |
V127 G2 |
V019 GM2 cell |
V031 GM2 cell |
V018 Kc167 cell |
V024 kc167 cell |
V073 mbn2 |
V148 mbn2 |
V075 ML-DmBG1-C1 |
V129 ML-DmBG1-c1 |
V076 ML-DmBG3-C2 |
V130 ML-DmBG3-c2 |
V034 ML-DmD16c3 cell |
V131 ML-DmD16-c3 |
V021 ML-DmD21 cell |
V010 MLDmD20c5 |
V036 ML-DmD20c5 cell |
V027 ML-DmD21 cell |
M005 ML-DmD32 cell |
M006 ML-DmD32 |
V022 ML-DmD32 cell |
V035 ML-DmD32 cell |
V132 ML-DmD32 |
V030 ML-DmD8 cell |
V134 ML-DmD8 |
V029 ML-DmD9 cell |
V125 ML-DmD9 |
M029 OSC cells (nuclear) |
M030 OSC cells (cytoplasmic) |
M031 OSC cells (whole cell) |
V144 OSC |
OSS2 OSS_s2 |
OSS6 OSS_s6 |
OSS7 OSS_s7 |
OSS8 OSS_s8 |
V032 S1 cell |
V146 S1 cell |
V008 S2-DRSC |
V145 S2-DRSC |
V067 S2-NP |
V020 S2R+ cell |
V025 S2R+ cell |
V074 S3 |
V128 S3 |
V011 Sg4 |
V133 Sg4 |
T004 OSC-4N |
T005 89-4N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGAGAAGGGTTTCGTGTG............................................................................................................... | 20 | 0 | 20 | 181.70 | 3634 | 3403 | 0 | 0 | 0 | 0 | 0 | 86 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 136 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGTGGAGAAGGGTTTCGTGTG............................................................................................................... | 21 | 0 | 20 | 36.95 | 739 | 0 | 139 | 0 | 0 | 0 | 75 | 0 | 0 | 407 | 0 | 0 | 0 | 0 | 0 | 0 | 112 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGTGGAGAAGGGTTTCGT.................................................................................................................. | 18 | 0 | 20 | 20.25 | 405 | 319 | 10 | 0 | 0 | 0 | 0 | 0 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 0 | 0 | 0 | 17 | 0 | 0 | 2 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| ..................................................GTGGAGAAGGGTTTCGTGTGAC............................................................................................................. | 22 | 0 | 1 | 20.00 | 20 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 9 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGTTGATCACGAGTTAGTCGGTCCT................................................................................. | 25 | 0 | 20 | 15.45 | 309 | 0 | 42 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 65 | 0 | 9 | 0 | 119 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGGAGAAGGGTTTCGTGTG............................................................................................................... | 19 | 0 | 20 | 13.50 | 270 | 0 | 0 | 0 | 0 | 0 | 51 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 42 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 136 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 |
| ............................................................................GTTGATCACGAGTTAGTCGGTCCT................................................................................. | 24 | 0 | 20 | 11.70 | 234 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 202 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGGAGAAGGGTTTCGTGT................................................................................................................ | 19 | 0 | 20 | 11.40 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 156 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| .....................................................GAGAAGGGTTTCGTGTGAC............................................................................................................. | 19 | 0 | 1 | 11.00 | 11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGGAGAAGGGTTTCGTG................................................................................................................. | 18 | 0 | 20 | 10.95 | 219 | 0 | 0 | 1 | 0 | 0 | 0 | 94 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 27 | 6 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 34 | 0 | 0 | 0 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 1 | 0 |
| .................................................AGTGGAGAAGGGTTTCGTGT................................................................................................................ | 20 | 0 | 20 | 10.80 | 216 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 67 | 137 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGGAGAAGGGTTTCGTGT................................................................................................................ | 18 | 0 | 20 | 7.95 | 159 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 88 | 0 | 0 | 0 | 0 | 2 | 0 | 4 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 0 | 54 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CACGAGTTAGTCGGTCCTAA............................................................................... | 20 | 0 | 20 | 7.75 | 155 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 0 | 82 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 9 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 1 | 0 |
| ...................................................TGGAGAAGGGTTTCGTGTGAC............................................................................................................. | 21 | 0 | 1 | 7.00 | 7 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................AGAAGGGTTTCGTGTGAC............................................................................................................. | 18 | 0 | 2 | 7.00 | 14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CACGAGTTAGTCGGTCCTAAG.............................................................................. | 21 | 0 | 20 | 5.55 | 111 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 9 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 74 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGTGGAGAAGGGTTTCGTGTGA.............................................................................................................. | 22 | 0 | 20 | 5.10 | 102 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 | 0 | 2 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| ...................................................................................ACGAGTTAGTCGGTCCTAA............................................................................... | 19 | 0 | 20 | 4.95 | 99 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 63 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGGAGAAGGGTTTCGTGTGA.............................................................................................................. | 21 | 0 | 20 | 4.85 | 97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 66 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGTTGATCACGAGTTAGTCGGTCC.................................................................................. | 24 | 0 | 20 | 4.75 | 95 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 51 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GTGGTTGATCACGAGTTAGTCGGTCCT................................................................................. | 27 | 0 | 20 | 4.75 | 95 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 45 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 11 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 7 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GTATCGTGTTCCAGTGGTTG..................................................................................................... | 20 | 3 | 6 | 4.50 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 18 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................GGTTGATCACGAGTTAGTCGGTCCTA................................................................................ | 26 | 0 | 20 | 4.35 | 87 | 51 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGGAGAAGGGTTTCGTGTGA.............................................................................................................. | 20 | 0 | 20 | 4.20 | 84 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 1 | 2 | 0 | 0 |
| ....................................................GGAGAAGGGTTTCGTGTG............................................................................................................... | 18 | 0 | 20 | 4.20 | 84 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CACGAGTTAGTCGGTCCTAAGT............................................................................. | 22 | 0 | 20 | 4.05 | 81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 71 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CACGAGTTAGTCGGTCCTA................................................................................ | 19 | 0 | 20 | 3.80 | 76 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 62 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................GAGAAGGGTTTCGTGTGA.............................................................................................................. | 18 | 0 | 20 | 3.75 | 75 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................ATCACGAGTTAGTCGGTCCT................................................................................. | 20 | 0 | 20 | 3.50 | 70 | 65 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CACGAGTTAGTCGGTCCT................................................................................. | 18 | 0 | 20 | 3.10 | 62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 |
| .............................................................................................................GCGAAAGCCGAAAATTTTCAA................................................... | 21 | 0 | 20 | 3.00 | 60 | 0 | 0 | 0 | 30 | 0 | 6 | 0 | 10 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ..................................................GTGGAGAAGGGTTTCGTGTGTC............................................................................................................. | 22 | 1 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TAAGTTCAAGGCGAAAGC................................................................ | 18 | 0 | 20 | 3.00 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TAGTCGGTCCTAAGTTCAAGGC...................................................................... | 22 | 0 | 20 | 3.00 | 60 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 16 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGTGGAGAAGGGTTTCGTGTGAC............................................................................................................. | 23 | 0 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................ATCACGAGTTAGTCGGTCCTAAG.............................................................................. | 23 | 0 | 20 | 2.95 | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGTTGATCACGAGTTAGTCG...................................................................................... | 20 | 0 | 20 | 2.75 | 55 | 12 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGGAGAAGGGCTTCGTGTG............................................................................................................... | 20 | 1 | 20 | 2.55 | 51 | 50 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TTCAAGGCGAAAGCCGAAAA.......................................................... | 20 | 0 | 20 | 2.35 | 47 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47 | 0 |
| ...................................................................................ACGAGTTAGTCGGTCCTAAGTTC........................................................................... | 23 | 0 | 20 | 2.30 | 46 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TCACGAGTTAGTCGGTCCTAAGT............................................................................. | 23 | 0 | 20 | 2.25 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GTGGTTGATCACGAGTTAGTCGGTCCTA................................................................................ | 28 | 0 | 20 | 2.20 | 44 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 31 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TCACGAGTTAGTCGGTCCT................................................................................. | 19 | 0 | 20 | 2.15 | 43 | 17 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
|
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGTGGAGAAGGGTTTCGTGTGACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCGAAAATTTTCAAGTAAAACAAAAATGCCTAACTATATAAACAAAGCGAATTATAATACACTTG
***********************************................((((...(((..(((..((((.(((......)))..))))..))).)))...))))........****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V085 CME W2 wing disc |
V135 CME W2 (wing disc line) |
V026 1182-4H cell |
V147 1182-4H cell |
V009 CMEL1 |
V126 CME L1 |
V028 CMEW1 Cl.8+ cell |
V033 CMEW1 Cl.8+ cell |
V091 fGS/OSS total |
V127 G2 |
V019 GM2 cell |
V031 GM2 cell |
V018 Kc167 cell |
V024 kc167 cell |
V073 mbn2 |
V148 mbn2 |
V075 ML-DmBG1-C1 |
V129 ML-DmBG1-c1 |
V076 ML-DmBG3-C2 |
V130 ML-DmBG3-c2 |
V034 ML-DmD16c3 cell |
V131 ML-DmD16-c3 |
V021 ML-DmD21 cell |
V010 MLDmD20c5 |
V036 ML-DmD20c5 cell |
V027 ML-DmD21 cell |
M005 ML-DmD32 cell |
M006 ML-DmD32 |
V022 ML-DmD32 cell |
V035 ML-DmD32 cell |
V132 ML-DmD32 |
V030 ML-DmD8 cell |
V134 ML-DmD8 |
V029 ML-DmD9 cell |
V125 ML-DmD9 |
M029 OSC cells (nuclear) |
M030 OSC cells (cytoplasmic) |
M031 OSC cells (whole cell) |
V144 OSC |
OSS2 OSS_s2 |
OSS6 OSS_s6 |
OSS7 OSS_s7 |
OSS8 OSS_s8 |
V032 S1 cell |
V146 S1 cell |
V008 S2-DRSC |
V145 S2-DRSC |
V067 S2-NP |
V020 S2R+ cell |
V025 S2R+ cell |
V074 S3 |
V128 S3 |
V011 Sg4 |
V133 Sg4 |
T004 OSC-4N |
T005 89-4N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GTATCGTGTTCCAGTGGTTG..................................................................................................... | 20 | 3 | 6 | 8.50 | 51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 45 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 |
| ...........................................................GGTATCGTGTTCCAGTGGTTG..................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TCGTGTGCCAGTGGTTGGTGA................................................................................................. | 21 | 3 | 6 | 0.50 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................CGTGTGCCAGTGGTTGGTGAC................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGAAAGCCGAAAATTTTCAAG.................................................. | 21 | 0 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................AGCCGAAAATTTTCAAGTAAA.............................................. | 21 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................AATATAAACAAAGCGAATTATA......... | 22 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................AAGCCGAAAATTTTCAAGTAA............................................... | 21 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GTATCGTGTTCCAGTGGTT...................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TGTATCGTGTGCCAGTGGTTG..................................................................................................... | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CCGAAAATTTTCAAGCAAAA............................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................CGAAAGCCGAAAATTTTCCAG.................................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGAAAGCCGAAAATTTTCACG.................................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................CAAAAATGGCTAACTATATAA........................ | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CCGAAAATTTTCAAGTAAAA............................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CCGAAAATTTTCGAGTAAAA............................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGTGGGTCCTAAATTCAAGGC...................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................CAGTGGTTGATCACGAGTTAG......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GGCGAAAGCCGAAAATTTTCA.................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................ATAAACAAAGCGAATTATAAT....... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CAGAAAATTTTCAAGTAAAAC............................................ | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ACAGTTTTTGATCACGAGTGA.......................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CGAAAACTTTCAAGTAAAACAAAAA....................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................TAAACAAAGCGAATTATAACA...... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGAAAGCCGAAAATTTTCAA................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AACAAAGCGAATTATAATACA.... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGAAAGCCGAAAATTTTCCAT.................................................. | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TGGTTGATCACGAGTCAGTC....................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................GCGAAAGCCGAAAATTTTCA.................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................CAAAAATGCCCAACCATATAA........................ | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AACGCCGAAAATTTTCAAGTA................................................ | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AAAAATGCCCAACCATATGAA....................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TAAAACAAAAATGCCTAACTA............................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | miRBase ID | Alignment |
|---|---|---|---|
| dm3 | chrUextra:25300070-25300250 + | NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGTGGAGAAGGGT-TTCGTGTGA-CAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACAAAAATGCC--------------------------TAAC-------TATATAA------------AC------AAA-GCGAATTATAATACACTTG | |
| droSim3 | chrU_M_4332:733-863 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGA-CAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACAAAAATGCC--------------------------TAAC-------TATATAA------------AC------AAA-GCGAA-TATAATACACTTG | |
| droSec2 | scaffold_4075:426-557 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACAAAAATGCC--------------------------TAAC-------TATATAA------------AC------AAA-GCGAA-TATAATACACTTG | |
| droYak3 | v2_chrUn_225:340-471 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACAAAAATGCC--------------------------AAAC-------TACATAA------------A-------AAA-GCGAATTATAATACACTTG | |
| droEre2 | scaffold_4512:190197-190328 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAATTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACAAAAATGCC--------------------------AAAT-------TATATAA------------AC------AAA-GCGAA-AATAATACACTTG | |
| droEug1 | scf7180000408050:587-713 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAAA-------CT--------------------------GAAA-------TATACAA------------AT-A----AAAGCTGGA-AATAATATACTTG | |
| droBia1 | scf7180000298283:164-308 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAATACAAATGCC--------------------------AAACAAATATATATATTA------------TATAAAATCTA-GCTAA-ATTAATATACTTG | |
| droTak1 | scf7180000411785:649-780 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACCAAAAACGC--------------------------AATA-------TATACAA------------AT------CAA-GCTAA-TATAATATACTTG | |
| droEle1 | scf7180000487776:706-851 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTTCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAACAAAAA-G-CAAACTATTAAGAAACACAAATAA-------------------TA------------AT-AA---AAG-GCAAA-AATAATATACTTG | |
| droRho1 | scf7180000764684:596-735 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCGAAAAATTTTCAAGTAAAACCAAAAAGCG--------------------------AATA-----TATATATAT------------AC-AAA-TAAT-GCCAATAATAATATACTTG | |
| droFic1 | scf7180000450076:840-975 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAAAACTAATGAC--------------------------TTAA---TAAATATATAT------------AC------AAG-CCAAA-AATAATATACTTG | |
| droAna3 | scaffold_8078:263-384 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAAATAAAAT-----------------------------TTAC-------TATAT--------------------------GTAAA-AATTATATACTTG | |
| droBip1 | scf7180000393153:5437-5560 - | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAAATAAATA-----------------------------------------TTACAA------------TT------ATT-GTGAA-AATTATATACTTG | |
| dp5 | Unknown_singleton_2130:123-257 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAAAATCAGTGAG--------------------------TAA----------TATTGTCAAATCTTTTA-------------TGAT-TTTTAAATACTTG | |
| droPer2 | scaffold_271:20398-20532 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAAAATCAGTGAG--------------------------TAA----------TATTGTCAAATCTTTTA-------------TGAT-TTTTAAATACTTG | |
| droWil2 | scf2_1100000003192:1711-1835 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTT--TCAAAAATGTC--------------------------AAAG-------GAAATAA------------T-------A------AATAATAAAACACTTG | |
| droVir3 | scaffold_5278:849-981 + | -------------------------------------------------AGTGGAGAAGGGT-TTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCG-AAAATTTTCAAGTAAAGA-AAAAT--C--------------------------TAAA---TATATTTATCA------------TC------TAA-CCGAT-TTTTATACACTTG | |
| droMoj3 | scaffold_6499:165666-165810 - | -------------------------------------------------AGTGGAGAAGGGTTTTCGTGTGAACAGTGGTTGATCACGAGTTAGTCGGTCCTAAGTTCAAGGCGAAAGCCGGAAAATTTTCAAGTAAAGA-AAAA--TCAAAATA--AATAAACAAAAATGTGCCTAAT-------TAT-----------------------------CGAT-TTTTATACACTTG |
Generated: 02/13/2013 at 05:25 PM