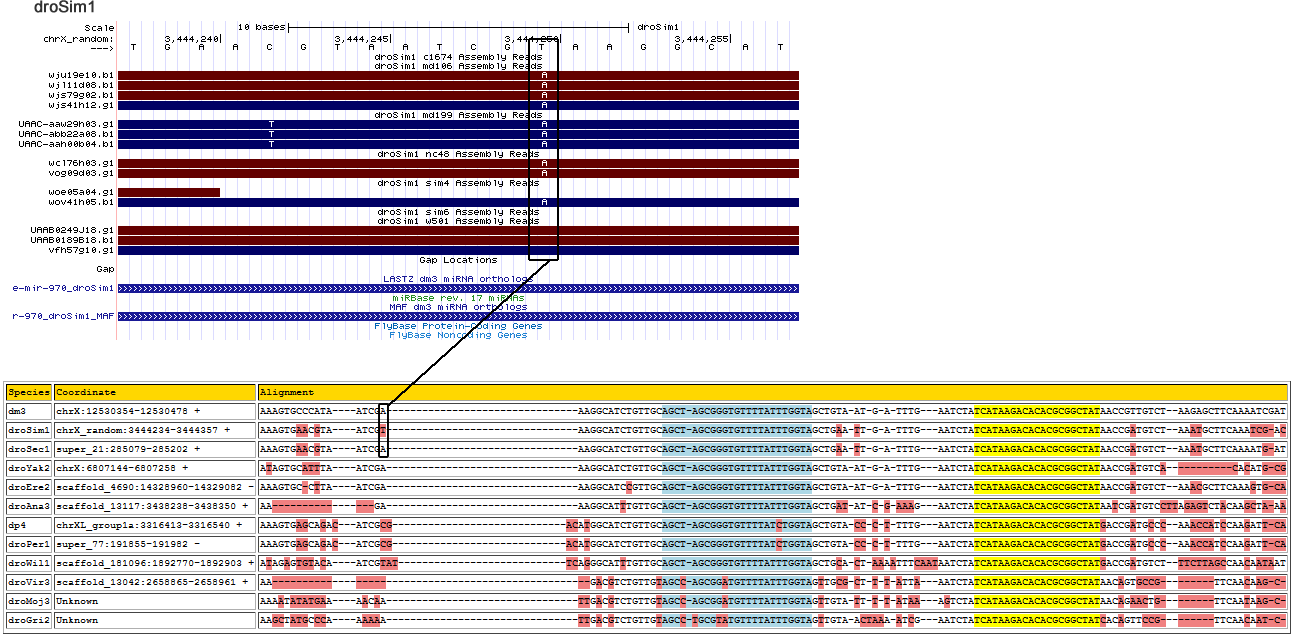
dme-mir-970
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chrX_random | 3444250 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0.769 | 0 | 0 | 0.231 | 13 | Genomic Error | 5' MOR |

Corrected Alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:12530354-12530478 + | AAAG-TGCCCAT-------------------AATCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGTAA-TG--ATTTG---AATCTATCATAAGACACACGCGGCTATAACCGTT--GTCT--AAGAGCTTCAA-----------A---------------ATCGAT |
| droSim1 | chrX_random:3444234-3444357 + | AAAG-TGAACGT-------------------AATCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGAAT-TG--ATTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCT--AAATGCTTCAA-----------A----------------TCGAC |
| droSec1 | super_21:285079-285202 + | AAAG-TGAACGT-------------------AATCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGAAT-TG--ATTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCT--AAATGCTTCAA-----------A----------------ATGAT |
| droYak2 | chrX:6807144-6807273 + | ATAG-TGCATTT-------------------AATCGAAAGGCATCTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGTAA-TG--ATTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCA--CACA--TGCGACACACACA-------------T------TAGAT |
| droEre2 | scaffold_4690:14328958-14329082 - | AAAG-TGCCTTA-------------------ATC-GAAAGGCATCCGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGTAA-TG--ATTTG---AATCTATCATAAGACACACGCGGCTATAACCGAT--GTCT--AAACGCTTCAA-----------AGT--------------GCAAT |
| droAna3 | scaffold_13117:3438231-3438350 + | TTAG-TGGAA-------------------------GAAAGGCATTTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGATA-TC--GAAAG---AATCTATCATAAGACACACGCGGCTATAATCGAT--GTCCTTAGAGTCTACAA---------------------------GCTAAA |
| dp4 | chrXL_group1a:3316403-3316556 + | ATCA-CCCCA--AAAAGTGAGCAGACATCG--CGA-CATGGCATCTGTTGCAGCTAGCGGGTGTTTTATCTGGTAGCTGTAC-CC--TTTTG---AATCTATCATAAGACACACGCGGCTATGACCGAT--GCCC--AAACCATCCAA-----------GATTCAATAATTAGTAATCAAT |
| droPer1 | super_77:191839-191992 - | ATCA-CCCCA--AAAAGTGAGCAGACATCG--CGA-CATGGCATCTGTTGCAGCTAGCGGGTGTTTTATCTGGTAGCTGTAC-CC--TTTTG---AATCTATCATAAGACACACGCGGCTATGACCGAT--GCCC--AAACCATCCAA-----------GATTCAATAATTAGTAATCAAT |
| droWil1 | scaffold_181096:1892772-1892900 + | AGAG-TGTAC------------A---ATCGTATTC--AGGGCATTTGTTGCAGCTAGCGGGTGTTTTATTTGGTAGCTGCAC-TAAAATTTCAATAATCTATCATAAGACACACGCGGCTATGACCGAT--GTCT--T------TC-----------------------TTAGCCAACAAT |
| droVir3 | scaffold_13042:2658865-2658967 + | ---------------------------------------GACGTCTGTTGTAGCCAGCGGATGTTTTATTTGGTAGTTGCGC-TT--TATTA---AATCTATCATAAGACACACGCGGCTATAACAGTGCCGTTC--AACA--AGCAAC-----------------------------GTC |
| droMoj3 | scaffold_6328:939989-940116 - | TAAA-CCCCAGT------GATCGACTATGA-AAACAATTGACGTCTGTTGTAGCCAGCGGATGTTTTATTTGGTAGTTGTAT-TT--TATAA---AGTCTATCATAAGACACACGCGGCTATAACAGAACTGTTC--AATA-------------------------------------AGC |
| droGri2 | scaffold_15203:5475247-5475378 + | AAAG-TCTGCCC-------------------AAAAAATTGACGTCTGTTGTAGCCTGCGTATGTTTTATTTGGTAGTTGTAACTAA-AATCG---AATCTATCATAAGACACACGCGGCTATCACAGTTCCGTTC--AACA--ATCAACGTAC-CA--------------------TTACA |