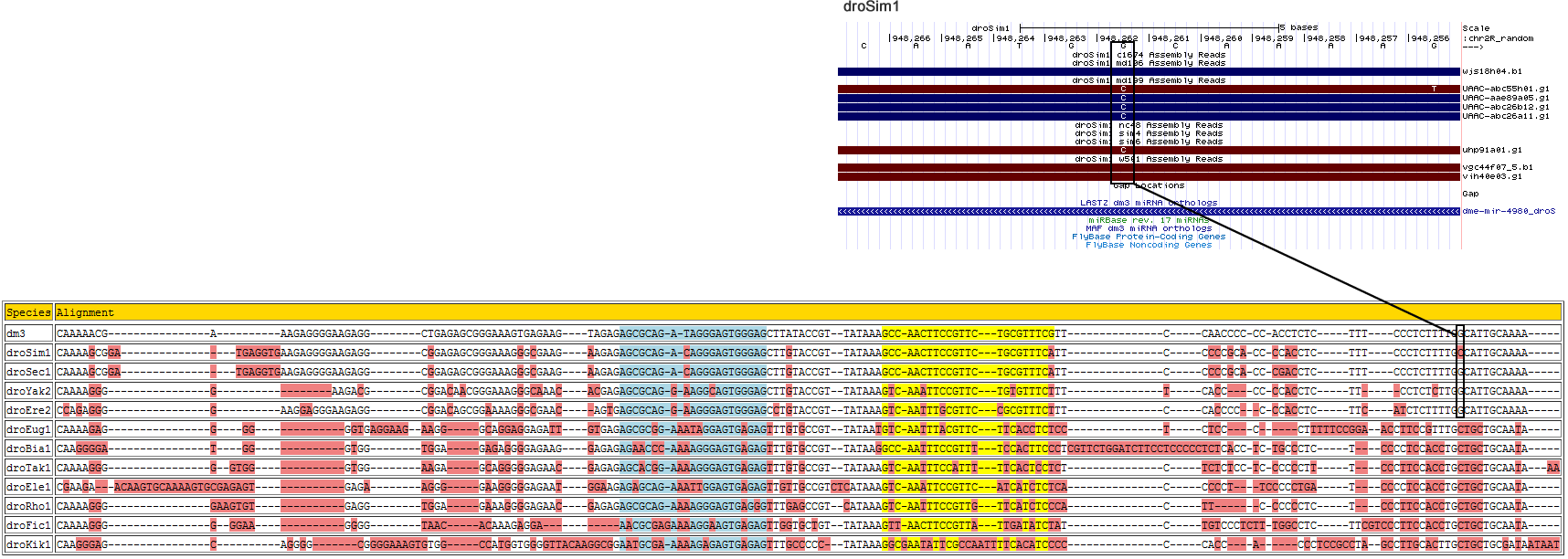
dme-mir-4980
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr2R_random | 948262 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.375 | 0.625 | 0 | 8 | Genomic Error | 3' flanking region |

Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:3673811-3673963 + | CAAAAA-CGAA--------------------AG-----AGGGGAAGAGGCTGAGAGCGGGAAAGT---GAGAAGT-----AGAGAGCGCAGA-TAGGGAGTGG----------------------------G----------------------AGCTTATACCGTT--ATAAAGCC-AACTTCCGTT----------------CTGCGTTTCGTTCCAAC-CC--C-------C-CACCTCTCTTT---------CCCTCTTTTGGCAT---------TGCAAAA----- |
| droSim1 | chr2R_random:948112-948272 + | CAAAAG-CGGA-TGA-----------GGTGAAG-----AGGGGAAGAGGCGGAGAGCGGGAAAGG---GCGAAGA-----AGAGAGCGCAGA-CAGGGAGTGG----------------------------G----------------------AGCTTGTACCGTT--ATAAAGCC-AACTTCCGTT----------------CTGCGTTTCATTCCCCC-GCACC-------C-CAC--CTCTTT---------CCCTCTTTTGGCAT---------TGCAAAA----- |
| droSec1 | super_1:1330268-1330428 + | CAAAAG-CGGA-TGA-----------GGTGAAG-----AGGGGAAGAGGCGGAGAGCGGGAAAGG---GCGAAGA-----AGAGAGCGCAGA-CAGGGAGTGG----------------------------G----------------------AGCTTGTACCGTT--ATAAAGCC-AACTTCCGTT----------------CTGCGTTTCATTCCCCC-GCACC-------C-GAC--CTCTTT---------CCCTCTTTTGGCAT---------TGCAAAA----- |
| droYak2 | chr2L:16346698-16346837 + | CAAAAG-GGGA--------------------AG-----ACG--------CGGACAACGGGAAAGG---GCAAACA-----CGAGAGCGCAGG-AAGGCAGTGG----------------------------G----------------------AGCTTGTACCGTT--ATAAAGTC-AAATTCCGTT----------------CTGTGTTTCTTTTCACC----CC-------C-CAC--CTCTT-----------CCTCTCTTGGCAT---------TGCAAAA----- |
| droEre2 | scaffold_4929:18933782-18933931 - | CCAGAG-GGGA--------------------AG-----GAGGGAAGAGGCGGACAGCGGAAAAGG---GCGAAC------AGTGAGCGCAGG-AAGGGAGTGG----------------------------G----------------------AGCCTGTACCGTT--ATAAAGTC-AATTTGCGTT----------------CCGCGTTTCTTTCCACC-CC--C-------C-CAC--CTCTTC---------ATCTCTTTTGGCAT---------TGCAAAA----- |
| droAna3 | scaffold_13266:10179298-10179443 + | AGCAGA-CGAG----------------GTGCAG-----GGGGAGAATAAAAATAACCAAAAAAAA---TAATAAG-----ATAAAGTGAACT-AAGAGAGTTG----------------------------G----------------------AATTT---CCATA--TCAGCACC-CATTTTCGAT----------------TTACATTCTCCTCCACC--------------------CTC----CC-----CACCCCACCTGCTGCTG---G---TGCAA------- |
| dp4 | chrXL_group1e:1273472-1273542 - | AGAGAG-AGAG--------------------AG-----AGAGAGAGAGAGAGAGAGAGGGAGAGG---GAGAGGG-----AGAGGGTGATGG-TTGGGTGTGG----------------------------G----------------------AG--------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 | super_4:2253896-2254021 + | CAAGAG-AGAG--------------------AG-----AGAGAGAGAGAAAGAGAGCGAGAAAGA---TAGAAGC-----AGAGCGAGA----GCGAAAGTGA----------------------------T--------------AGATATATAGTTTATACC-------AAACTT-TTTTAATAAT----------------CCATGTTCTTTTCG-------------------AT--GTT----TA-----TTTTC------------------------------- |
| droWil1 | scaffold_181096:5834838-5834931 + | AGAGAC-AGAG--------------------AG-----AGAGAGAGAGAGAGAGAGAGAGAGACA---GAAGAGT-----GGTGAGTGGTGG-AAGGGGGTGG----------------------------G----------------------GGCCAATATTGTG--CTAGTGCT-GAAT------------------------------------------------------------------------------------------------------------- |
| droVir3 | scaffold_12963:19208738-19208883 + | TTAGTA-AGAG--------------------AG-----AGAGAGAGAGAGAGGGAGAGCCAGCGA---GTAA--T-----AGAGAG----------CACATGC----------------------------G----------------------AGCATAAACCGGT--TTGAG--C-GCTCTCCGCT-------GGCTAT---GCGCGCTCCCAGGCCAG-C--GCTCACCTGTG-TA--CAAGTT---------CACTTTGTTGTTGT---------TG---------- |
| droMoj3 | scaffold_6496:13778911-13779004 + | CAACAG-AGAC--------------------AG-----AGAGAGAGAGAGAGAGAGCGGGAGAGA---GAGAAAG-----AGAGAGAGA----GAGAGAGGGA----------------------------G-----------------------GCCATGGCTGTA--GCTGAGCT-GCT-TACGT-------------------------------------------------------------------------------------------------------- |
| droGri2 | scaffold_15110:15752678-15752830 - | AGTGAG-AGAG--------------------AG-----AGAGAGAGAGAGAGAGAGAGAGAGAGA---GAG----------------------AGAGAGCTGGGCACTTGTGTCTGGGGCTGTGCTCATGCAAAAATAAATGCGAAC---------TGTGTGCAG-ATCGTAAATTA-----------------------------AAGTTTCTCTCGCTC-CC--------------C--CTATTT--------CTCTGTTCCTGT------------T----------- |