dme-mir-3641
Changes
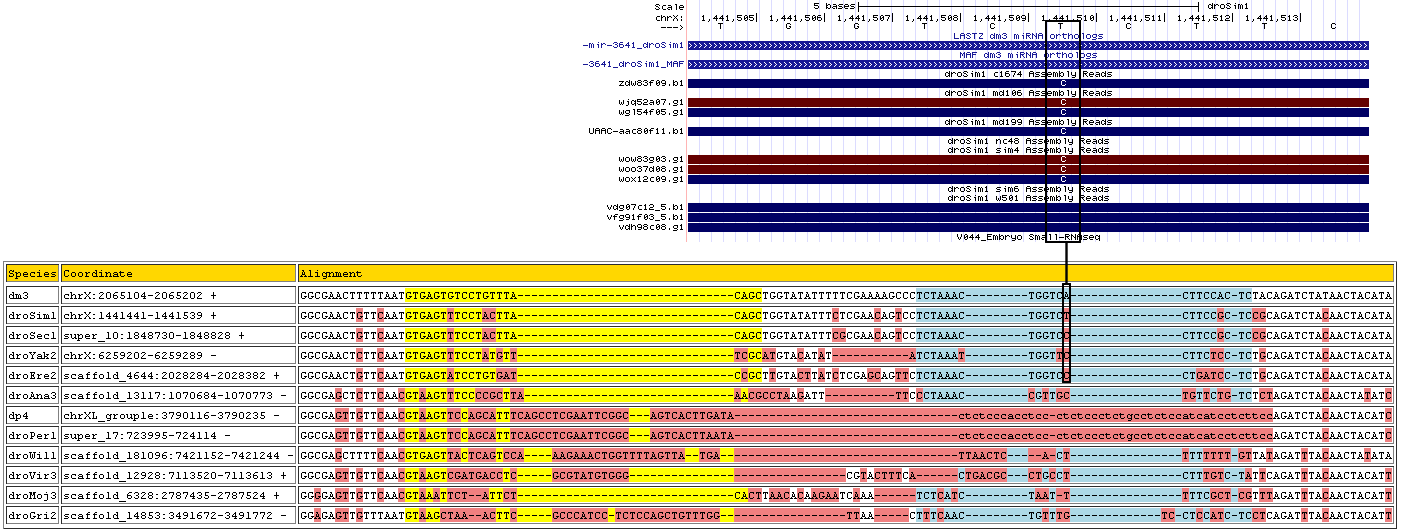
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chrX | 1441510 | T | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.7 | 0 | 0.3 | 10 | Genomic Error | miR* |

Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:2065104-2065202 + | GGCGAACTTTTTAATGTGAGTGTCCT-------G--------------------------------------------------------------------TTTACAGCTGGTATATTTTTCGAAAAGCCCTCTA-----AACTGGT-CACTTCCACTCTACAGATCTATAACTACATA |
| droSim1 | chrX:1441441-1441539 + | GGCGAACTGTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------CTTACAGCTGGTATATTTCTCGAACAGTCCTCTA-----AACTGGT-CCCTTCCGCTCCGCAGATCTACAACTACATA |
| droSec1 | super_10:1848730-1848828 + | GGCGAACTGTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------CTTACAGCTGGTATATTTCGCGAACAGTCCTCTA-----AACTGGT-CCCTTCCGCTCCGCAGATCTACAACTACATA |
| droYak2 | chrX:6259202-6259289 - | GGCGAACTCTTCAATGTGAGTTTCCT-------A--------------------------------------------------------------------TGTTTCGCATGTACATA-----------TATCTA-----AATTGGT-TCCTTCTCCTCTGCAGATCTACAACTACATA |
| droEre2 | scaffold_4644:2028284-2028382 + | GGCGAACTGTTCAATGTGAGTATCCT-------G--------------------------------------------------------------------TGATCCGCTTGTACTTATCTCGAGCAGTTCTCTA-----AACTGGT-CCCTGATCCTCTGCAGATCTACAACTACATA |
| droAna3 | scaffold_13117:1070684-1070773 - | GGCGAGCTCTTCAACGTAAGTTTCCCCG------C--------------------------------------------------------------------TTAAACGCCTAA----------GATTTTCCCTA-----AACCGTT-GCTGTTCTGTCTCTAGATCTACAACTATATC |
| dp4 | chrXL_group1e:3790116-3790235 - | GGCGAGTTGTTCAACGTAAGTTCCAGCATTTCA-GCCTCGAATTCGGCAGTCACTTGA------------------------TACTCTCCCACCTCCCTCTC--------------------CCTC-------------TG--CCTCTCCATCATCCTCTTCCAGATCTACAACTACATC |
| droPer1 | super_17:723995-724114 - | GGCGAGTTGTTCAACGTAAGTTCCAGCATTTCA-GCCTCGAATTCGGCAGTCACTTAA------------------------TACTCTCCCACCTCCCTCTC--------------------CCTC-------------TG--CCTCTCCATCATCCTCTTCCAGATCTACAACTACATC |
| droWil1 | scaffold_181096:7421152-7421244 - | GGCGAGCTTTTCAACGTGAGTTACTCAG------T------------------------CCAAAGAAACTGGTTTTAGTTATGAT-----------------------------------------TAA--------------CTCA--CTTTTTTTTGTTATAGATTTACAACTATATA |
| droVir3 | scaffold_12928:7113520-7113613 + | GGCGAGTTGTTCAACGTAAGTCGATG-------A--------------------------------------------------------------------CCTCGCGTATGTGGG-----CGTACTTTCACTGA-----CGCCTGC-CTCTTTGTCTATTCAGATTTACAACTACATT |
| droMoj3 | scaffold_6328:2787435-2787524 + | GGGGAGTTGTTCAACGTAAATTCTAT-----------------------------------------------------------------------------TCTCACTTAACACAAGAATC----AAA--TCTC-----ATCTAAT--TTTTCGCTCGTTTAGATTTACAACTACATT |
| droGri2 | scaffold_14853:3491672-3491772 - | GGAGAGTTGTTTAATGTAAGCTAAA---------C-------TTCGCCCATCCT-------------------------------------------------CTCCAGCTGTTTGG-----TTAA-----CTTTCAACTG--TTTGT--CCTCCATCTCCTCAGATTTACAACTACATT |