dme-mir-306

The correct ortholog was found in its entirety in 6 of the 7 strains, and partially found in md106.
>dme-mir-306-dm3
GTGAATAGTTTAAAAGTCCACTCGATGGCTCAGGTACTTAGTGACTCTCAATGCTTTTGACATTTTGGGGGTCACTCTGTGCCTGTGCTGCCAGTGGGACATAATCTACAAATAA
>zdv96e12.b1:525-640
GTGAATAGTTTAAAAGTCCACTCGATGGCTCAGGTACTTAGTGACTCTCAATGCCTTTGACATTTTGGGGGTCACTCTGTGCCTGTGCTGCCAGTGGGACATAATCTACAAATAA
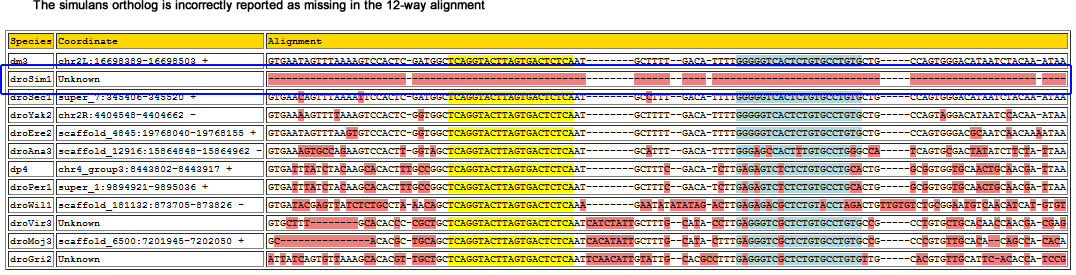
This miRNA lie in the same intron of the grp gene as 79 and 9b, and one intron downstream of 9c. The genome browser net track shows that this region's alignment with simulans is ambiguous:
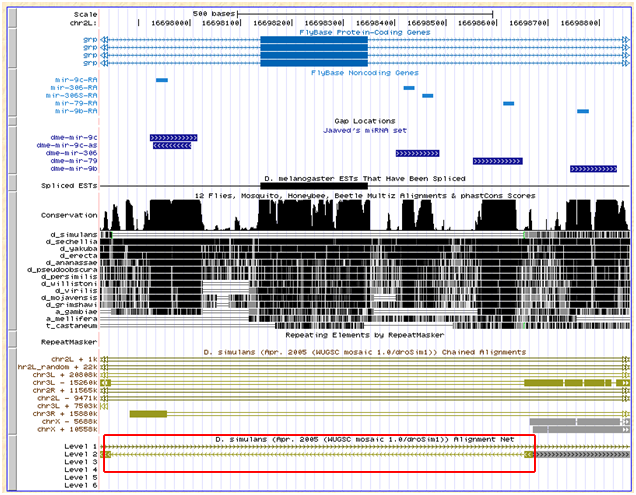
Flybase also does not report an ortholog for grp in simulans, however one exists for sechelia (GM17194).
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:16698389-16698503 + | GTGAATAG-TTTAAA------AG--------------TCCACTC-G-----ATGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACATA-ATCTACA----AATA-------------A |
| droSim1 | zdv96e12.b1:525-640 + | GTGAATAG-TTTAAA------AG--------------TCCACTC-G-----ATGGCTCAGGTACTTAGTGACTCTCAAT--------GCCTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACATA-ATCTACA----AATA-------------A |
| droSec1 | super_7:345406-345520 + | GTGAACAG-TTTAAA------AT--------------TCCACTC-G-----ATGGCTCAGGTACTTAGTGACTCTCAAT--------GCCTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACATA-ATCTACA----AATA-------------A |
| droYak2 | chr2R:4404548-4404662 - | GTGAAAAG-TTTTAA------AG--------------TCCACTC-G-----GTGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTAGGACATA-ATCCACA----AATA-------------A |
| droEre2 | scaffold_4845:19768040-19768155 + | GTGAATAG-TTTAAG------TG--------------TCCACTC-G-----GTGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTTGA--CATTTTGGGGGTCACTCTGTGCCTGTGCTG-----CCAGTGGGACGCA-ATCAACA---AAATA-------------A |
| droAna3 | scaffold_12916:15864848-15864962 - | GTGAAAGT-GCCAGA------AG--------------TCCACTT-G-----GTAGCTCAGGTACTTAGTGACTCTCAAT--------GCATTTGA--CATTTTGGGAGCCACTTTGTGCCTGGGCCA-----TCAGTGCGACTAT-ATCTTCT----ATTA-------------A |
| dp4 | chr4_group3:8443802-8443917 + | GTGATTTA-TCTACA------AG--------------CACACTTTG-----CCGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTCGA--CATCTTGAGAGTCTCTCTGTGCCTGCACTG-----GCGGTGGTGCAAC-TGCAACG----ATTA-------------A |
| droPer1 | super_1:9894921-9895036 + | GTGATTTA-TCTACA------AG--------------CACACTTTG-----CCGGCTCAGGTACTTAGTGACTCTCAAT--------GCTTTCGA--CATCTTGAGAGTCTCTCTGTGCCTGCACTG-----GCGGTGGTGCAAC-TGCAACG----ATTA-------------A |
| droWil1 | scaffold_181132:873705-873839 - | GTGACTATACAAAGTGATACGAG--------------TTATCTCTGCCTAAACAGCTCAGGTACTTAGTGACTCTCAAA--------GAATATATATAGACTTGAGAGACGCTCTGTACCTAGACTGTTGTGTCTGCGGAATGTC-AACATCA----TGTG-------------T |
| droVir3 | scaffold_12963:12863192-12863320 - | GTGCTG--------A------ACTC-CTCCCGCCCTGCACACCC-C-----GCTGCTCAGGTACTTAGTGACTCTCAATCATCTATTGCTTTGCA--TACCTTGAGGGTCGCTCTGTGCCTGTGCCG-----CCTGTGCTGCACA-ACCAACG----ACGA-------------G |
| droMoj3 | scaffold_6500:7201931-7202062 + | ATTCATGA-TGTCC-------AG--------------CACACGC-T-----GCAGCTCAGGTACTTAGTGACTCTCAATCACATATTGCTTTGCA--TACTTTGAGGGTCGCTCTGTGCCTGTGCCG-----CCCGTGTTGCACACAGCCACA----CA---ATTGTGA-ATTCG |
| droGri2 | scaffold_14978:830883-831041 + | GTGTTAAA-TGTGGAAAAACCAATCCAATCCG--CTGCACACGT-T-----GCTGCTCAGGTACTTAGTGACTCTCAATTCAACATTGTATTGCA-CGCCTTTGAGGGTCGCTCTGTGCCTGTGTTG-----CACGTGTTGCATT-CACACCATCCGAGTGATTTGTGTCCTTTG |