dme-mir-285
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droSim1 | chr3L | 11308082 | C | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.278 | 0 | 0.722 | 18 | Genomic Error | 3' flanking region |
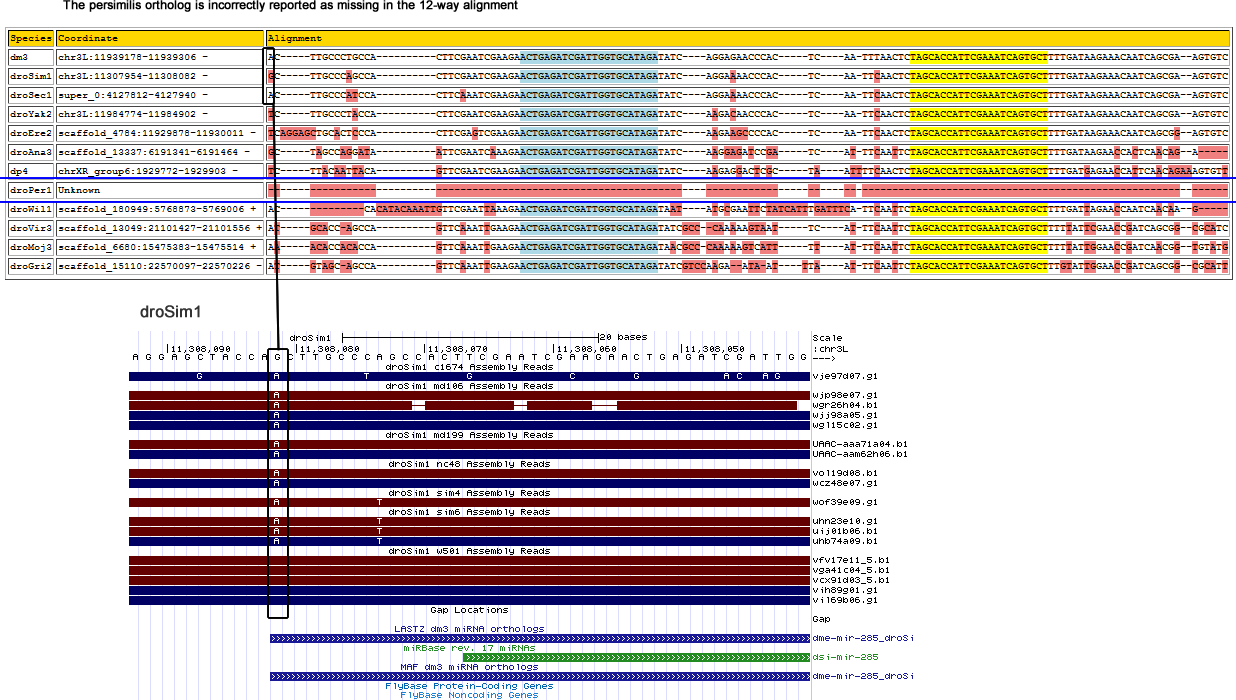
The reported persimilis ortholog reported in the 12-way alignment is wrong as shown below:

This miRNA is intergenic and there appear to be no nearby genes. The correct persimilis ortholog is found by searching the persimilis trace reads for the pseudobscura sequence via LASTZ.
>dme-mir-285-dp4
TCTTACAAT-TACAGTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATCAAGAGGACTCGCTAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA
>gnl|ti|738578230:636-763
TCTTACAATTTACAGTGGGAATCGAAGAACCTGAGATCGATTGGTGCATAGATATCAAGAGGACTCGATAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:11939178-11939306 - | AC--TT-------------------------GCCC-TGCCACTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAGGAGA---ACCCA-----CTCAA-------TTTAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAAG------------TGTC- |
| droSim1 | chr3L:11307954-11308082 - | AC--TT-------------------------GCCC-AGCCACTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAGGAAA---ACCCA-----CTCAA-------TTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAAG------------TGTC- |
| droSec1 | super_0:4127812-4127940 - | AC--TT-------------------------GCCC-ATCCACTTCAAATCGAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAGGAAA---ACCCA-----CTCAA-------TTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAAG------------TGTC- |
| droYak2 | chr3L:11984774-11984902 - | TC--TT-------------------------GCCC-TACCACTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAAGACA---ACCCA-----CTCAA-------TTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGAAG------------TGTC- |
| droEre2 | scaffold_4784:11929878-11930011 - | TCA--------GGAGCT--------------GCAC-TCCCACTTCGAGTCGAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAAGAAG---CCCCA-----CTCAA-------TTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAAACAATCAGCGGAG------------TGTC- |
| droAna3 | scaffold_13337:6191341-6191461 - | GC-------------------------------CA-GGATAATTCGAATCAAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAAGGAG---ATCCG-----ATCAT-------TTCAATTCTAGCACCATTCGAAATCAGTGCTTTTGATAAGAACCACTCAACAGA------------------ |
| dp4 | chrXR_group6:1929779-1929903 - | TC--TT-------------------------ACAA-TTACAGTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATAT-----CAAGAGG---ACTCG-----CTAATT------TTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA------------------ |
| droPer1 | gnl|ti|738578230:636-763 + | TC--TT-------------------------ACAATTTACAGTGGGAATCGAAGAACCTGAGATCGATTGGTGCATAGATAT-----CAAGAGG---ACTCG-----ATAATT------TTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA------------------ |
| droWil1 | scaffold_180949:5768880-5769006 + | ACAA-------------------------------A---TTGTTCGAATTAAAGAAC-TGAGATCGATTGGTGCATAGATAA-----TATGCGA---ATTCTATCATTT---GATTTCATTCAATTCTAGCACCATTCGAAATCAGTGCTTTTGATTAGAACCAATCAACAAG------------------ |
| droVir3 | scaffold_13049:21101427-21101556 + | AT--GCAC--------------------------C-AGCCAGTTCAAATTGAAGAAC-TGAGATCGATTGGTGCATAGATATCGCCC-A--AAA---AGTAA-----TTCAT-------TTCAATTCTAGCACCATTCGAAATCAGTGCTTTTTATTCGAACCGATCAGCGGCG------------CATC- |
| droMoj3 | scaffold_6680:15475365-15475526 + | AC--ACACCCAGCAGA-CATCAAACACCACA-------CCAGTTCAAATTGAAGAAC-TGAGATCGATTGGTGCATAGATAACGCCC-AA----AAA-----GTCATTTTAT-------TTCAATTCTAGCACCATTCGAAATCAGTGCTTTTTATTGGAACCGATCAACGGTGTATGGGAACAAGTGTC- |
| droGri2 | scaffold_15110:22570097-22570226 - | AT--GTA----GCA-----------------------GCCAGTTCAAATTGAAGAAC-TGAGATCGATTGGTGCATAGATATCGTCC-AAGAA----------TAATTTAAT-------TTCAATTCTAGCACCATTCGAAATCAGTGCTTTGTATTGGAACCGATCAGCGGCG------------CATT- |