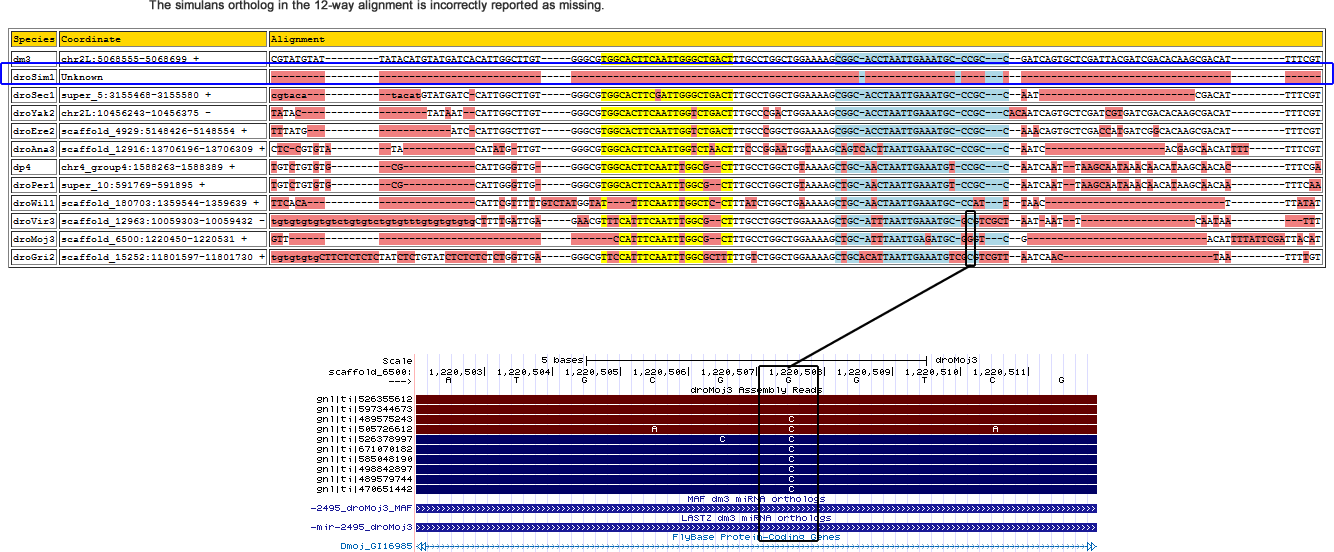
dme-mir-2495
Changes
| Species | Chrom | Pos | Ref base | Type | A | C | G | T | RNAseq Total | A | C | G | T | Trace Total | Decision | Partition |
| droMoj3 | scaffold_6500 | 1220508 | G | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.8 | 0.2 | 0 | 10 | Genomic Error | miR* |

>dme-mir-2495-dm3
CGTATGTATTATACATGTATGATCACATTGGCTTGTGGGCGTGGCACTTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCACCTAATTGAAATGCCCGCCGATCAGTGCTCGATTACGATCGACACAAGCGACATTTTCGT-----------------
>zds89d09.b1:77-211
CATACGTAC-ATACATGTATGATC-CATTGGCTTGTGGGCGCGGCACTTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCACCTAATTGAAATGCCCGCCAAT--------------------------CGACATTTTCGTTTAATTTCGTTTCTAGC
The D. simulans ortholog was only found in 3 of the 7 strains and not in w501. This D. melanogaster miRNA lies in an intron of the gene qtc. The genome browser indicate that the simulans alignment in this area is ambiguous as evident from chains in the browser screenshot below. As it appears, the ortholog for qtc would lie in an unlikely rearrangement event among chrU, chr3R, and chr2L_random:
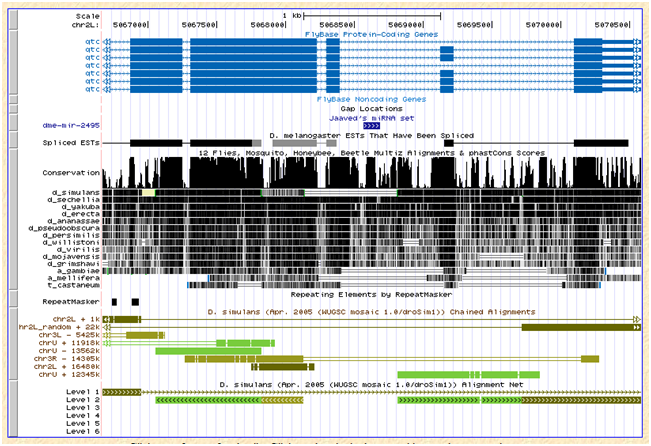
Flybase reports an ortholog of dsim\GD23320 for qtc, but this annotation was recently withdrawn. No ortholog is reported for sechelia but for D. erecta and D. yakuba.
Corrected alignment
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5068555-5068699 + | CGTAT--GTATT-------ATACATG----TA-T-------------------------GATC---AC-----A-T-TGGCTTGTGGGCGTGG--CAC-TTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGG-CAC-CTAATTGAAATGC-CC-GC---CGAT------CA---GTGCTCGAT-----------TAC---GATC----GACACAAGCGACATTTT---CGT----------------- |
| droSim1 | zds89d09.b1:77-211 + | CATAC--GTAC--------ATACATG----TA-T-------------------------GATC----C-----A-T-TGGCTTGTGGGCGCGG--CAC-TTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGG-CAC-CTAATTGAAATGC-CC-GC---CAAT-----------------------------------------------------CGACATTTT---CGTTTAATTTCGTTTCTAGC |
| droSec1 | super_5:3155468-3155580 + | CGTAC--------------ATACATG----TA-T-------------------------GATC----C-----A-T-TGGCTTGTGGGCGTGG--CAC-TTCGATTGGGCTGACTTTGCCTGGCTGGAAAAGCGG-CAC-CTAATTGAAATGC-CC-GC---CAAT-----------------------------------------------------CGACATTTT---CGT----------------- |
| droYak2 | chr2L:10456243-10456376 - | TATAC-------------------------TA-T-------------------------AAT-----C-----A-T-TGGCTTGTGGGCGTGG--CAC-TTCAATTGGTCTGACTTTGCCCGACTGGAAAAGCGG-CAC-CTAATTGAAATGC-CC-GC---CA--CAA--TCA---GTGCTCGAT-----------CGT---GATC----GACACAAGCGACATTTT---CGT----------------- |
| droEre2 | scaffold_4929:5148425-5148554 + | TTTA-----------------------------T-------------------------GATC----C-----A-T-TGGCTTGTGGGCGTGG--CAC-TTCAATTGGTCTGACTTTGCCCGGCTGGAAAAGCGG-CAC-CTAATTGAAATGC-CC-GC---CAAA------CA---GTGCTCGAC-----------CAT---GATC----GGCACAAGCGACATTTT---CGT----------------- |
| droAna3 | scaffold_12916:13706207-13706309 + | CATAT--G-------------------------------------------------------------------------TTGTGGGCGTGG--CAC-TTCAATTGGTCTAACTTTCCCGGAATGGTAAAGCAGTCAC-TTAATTGAAATGC-CC-GC---CAAT------C-----------------------------------------ACGAGCAACATTTTTTTCGT----------------- |
| dp4 | chr4_group4:1588263-1588389 + | TGTCT--GT-----------------------GTGCG------------------------------C-----A-T-TGGGTTG-GGGCGTGG--CAC-TTCAATTTGGC--GCTTTGCCTGGCTGTAAAAGCTG-CAA-CTAATTGAAATGT-CC-GC---CAAT------CA---ATTAA--GC-----------AAT---AAAC----AACATAAGCAACACTTT---CGA----------------- |
| droPer1 | super_10:591769-591895 + | TGTCT--GT-----------------------GTGCG------------------------------C-----A-T-TGGGTTG-GGGCGTGG--CAC-TTCAATTTGGC--GCTTTGCCTGGCTGTAAAAGCTG-CAA-CTAATTGAAATGT-CC-GC---CAAT------CA---ATTAA--GC-----------AAT---AAAC----AACATAAGCAACAATTT---CAA----------------- |
| droWil1 | scaffold_180703:1359544-1359639 + | TTCAC--A-----------------------------------------------------------C-----A-T-TCGTTTTTGTCTATGG--TATTTTCAATTTGGCTC-CTTTATCTGGCTGAAAAAGCTG-CAA-CTAATTGAAATGC-CC-AT---T-----------------------TAACT----------------------------------TT---ATAT----------------- |
| droVir3 | scaffold_12963:10059299-10059444 - | CGTAT--GTGT--------GTGTGTGTGTG-------TGTCTGTGTCTGTGTTTGTGTG----TGTGC-----T-T-TTGATTGAGAACGTTT--CAT-TTCAATTTGGC--GCTTTGCCTGGCTGGAAAAGCTG-CAT-TTAATTGAAATGC-GC-GTCGCTAAT-AATT-CAA-----------TAATTT------------------------------------AT-----------TA-------- |
| droMoj3 | scaffold_6500:1220428-1220559 + | TTTGA--GT--------------------------A--------------------------T----C-----AT------TTTAGGGCGTGTTCCAT-TTCAATTTGGC--GCTTTGCCTGGCTGGAAAAGCTG-CAT-TTAATTGAGATGC-GC-GT---CGA-CAT--------TTTAT--TC-----GATTACA--TTT-AG-TTTAATTACAAGAAATATTTA---AGC----------------- |
| droGri2 | scaffold_15252:11801593-11801729 + | TGTGT--GTGT-----------------------GTGCTTCTCTCTCTATCTCTGTATC----TCTC---T-C---TCTGGTTGAGGGCGTTC--CAT-TTCAATTTGGCGCTTTTTGTCTGGCTGGAAAAGCTG-CACATTAATTGAAATGTCG-CGTCGTTAAT------CAAC----------TAATT-------------------------------------------------TTG-------- |