dme-mir-1016
No Changes

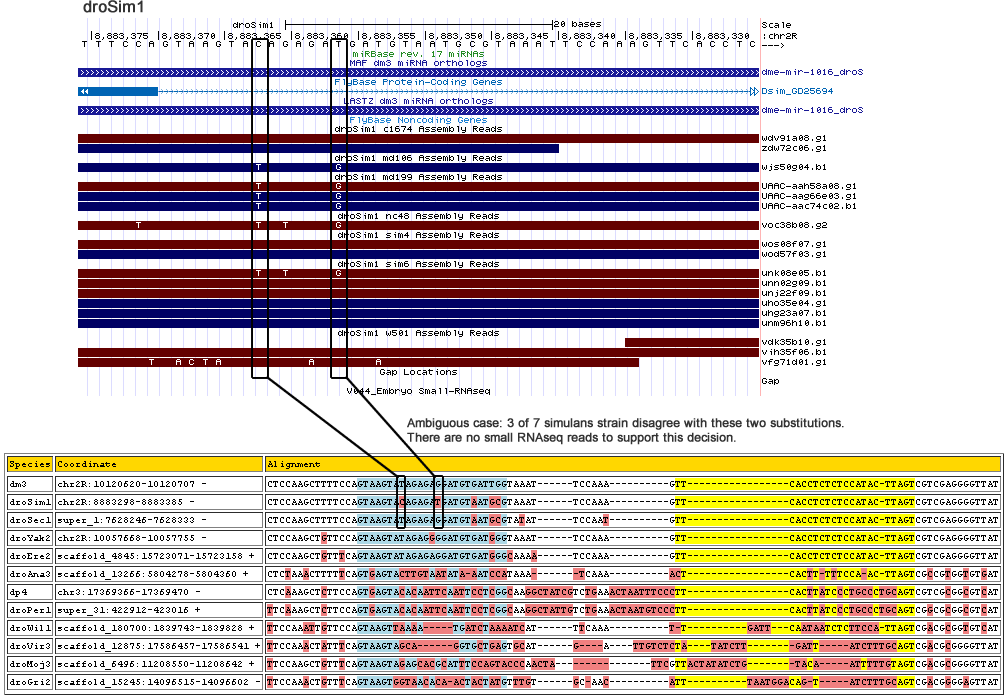
Uncorrected alignment:
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:10120620-10120707 - | CTCCAAGCTTTTCCAGTAAGTATAGAG---AG----GATGTGA-TT---GGTA-----------------AA-T---------------TCCAAAGTTCACCTCTCTCCA--TA-CTTAGTCGTCGAGGGGTTAT |
| droSim1 | chr2R:8883298-8883385 - | CTCCAAGCTTTTCCAGTAAGTACAGAG---AT----GATGTAA-TG---CGTA-----------------AA-T---------------TCCAAAGTTCACCTCTCTCCA--TA-CTTAGTCGTCGAGGGGTTAT |
| droSec1 | super_1:7628246-7628333 - | CTCCAAGCTTTTCCAGTAAGTATAGAG---AG----GATGTAA-TG---CGTA-----------------TA-T---------------TCCAATGTTCACCTCTCTCCA--TA-CTTAGTCGTCGAGGGGTTAT |
| droYak2 | chr2R:10057668-10057755 - | CTCCAAGCTGTTCCAGTAAGTATAGAG---GG----GATGTGA-TG---GGTA-----------------AA-T---------------TCCAAAGTTCACCTCTCTCCA--TA-CTTAGTCGTCGAGGGGTTAT |
| droEre2 | scaffold_4845:15723071-15723158 + | CTCCAAGCTGTTTCAGTAAGTATAGAG---AG----GATGTGA-TG---GGCA-----------------AA-A---------------TCCAAAGTTCACCTCTCTCCA--TA-CTTAGTCGTCGAGGGGTTAT |
| droAna3 | scaffold_13266:5804278-5804360 + | CTCTAAACTTTTTCAGTGAGTACTTGT---AA------TATAAATC---CATA-----------------A------------------ATCAAAACTCACTTT-TTCCA---A-CTTAGTCGCCGTGGTGTGAT |
| dp4 | chr3:17369366-17369470 - | CTCAAAGCTCTTCCAGTGAGTACACAA---TT----CAATTCC-TC---GGCAAGGCTATCGTCTGAAAC----TAATT----------TC---CCTTCACTTATCCCTG--CCCTGCAGTCGTCGCGGCGTCAT |
| droPer1 | super_31:422912-423016 + | TTCAAAGCTCTTCCAGTGAGTACACAA---TT----CAATTCC-TC---GGCAAGGCTATTGTCTGAAAC----TAATG----------TC---CCTTCACTTATCCCTG--CCCTGCAGTCGGCGCGGCGTCAT |
| droWil1 | scaffold_180700:1839743-1839828 + | TTCCAAATTGTTCCAGTAAGTTAAAAT---GA------T---C-TA---AAAT-----------------CATTTC------AA-----ATT--GATTCAATAATCTCTT--CC-ATTAGTCGACGCGGTGTCAT |
| droVir3 | scaffold_12875:17586457-17586541 + | TTCCAAACTATTTCAGTAAGTAGCAGG---T----------GC-TG---AGTG-----------------CA-TGATTGTC--------TCTATATCTTGATTATC------T-TTGCAGTCGACGCGGGGTTAT |
| droMoj3 | scaffold_6496:11208550-11208642 + | TTCCAAGCTGTTTCAGTAAGTAGAGCA---CG----CATTTCC-AG---TACCCAACTAT----------------TCGTT--------ACTATATCTGTACAATT------T-TTGTAGTCGACGCGGGGTTAT |
| droGri2 | scaffold_15245:14096515-14096602 - | TTCCAAACTGTTTCAGTAAGTGGTAAC---ACAACTACTATGT-TT-------------------------G-TGC-----AACA----TTTAATG-------GACAGTATCT-TTGCAGTCGACGGGGAGTTAT |