miRNA Name |
Query Species |
Result |
Experiment Needed |
Experiment Decision |
Sequence |
dme-mir-2282 |
Dm3 |
Found |
N |
NA |
>dme-mir-2882-dm3
TGGGGCAGTGGAGCTGCCTGCATAACGCTGTGGCTTTCCGCTTCTATTTTTCGTTCACTGCCTTGTGC----TTCGTTGGAAAATCGGTGAGCTAAAAATAGAATTCGGTTGCCACGCTTCAAGGGAAATGAGGAAAATTA
>zdo07f06.g1:327-468
TGGGGCAGTGGAGCTGCCTGCATAAGGCTGTGGCTTTCCGCTTCTATTTTTCGTTCACTGCCTTGTGCTTGCTTCGTTGGAAAATCGGTGAGCTAAAAATAGATTTCGGTTGCCACGCTTCAAGGGAAATGAGGAAAATTA
These ortholog lie in a gap of the simulans assembly (chr3L:2949116-2949968). The sequence was not found in the w501 strain's shotgun reads which comprises the bulk of the droSim1 assembly. It was found in the 6 other strains. |
dme-mir-2283 |
Dm3 |
Found |
N |
NA |
>dme-mir-2283-dm3
GAAAACTCACTCCTGAGCTGCTCCGAAAATATCA-----------TGAATACGACAATTATGTGCACATCATTTAGTATACGTGATATTTTAGGAGCAGCTAAATTATATTTAAAGT
>wcy63a10.b1:18-120
---------------AGTTGTTCCGAAAAAATCAATTTTAATGACAGGATGGGCAGATAATTGGGACATCATGTTGTATACGTAATATTTTAGGAGCAGCTCAATTATATTTAAAGT
This ortholog should lie in a region of the genome filled by a gap (chr3R:15989680-15990054). The ortholog was only found in the nc48 strain and in none of the other 6 strains. |
dme-mir-2495 |
Dm3 |
Found |
N |
NA |
>dme-mir-2495-dm3
CGTATGTATTATACATGTATGATCACATTGGCTTGTGGGCGTGGCACTTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCACCTAATTGAAATGCCCGCCGATCAGTGCTCGATTACGATCGACACAAGCGACATTTTCGT-----------------
>zds89d09.b1:77-211
CATACGTAC-ATACATGTATGATC-CATTGGCTTGTGGGCGCGGCACTTCAATTGGGCTGACTTTGCCTGGCTGGAAAAGCGGCACCTAATTGAAATGCCCGCCAAT--------------------------CGACATTTTCGTTTAATTTCGTTTCTAGC
The ortholog was only found in 3 of the 7 strains and not in w501. This dme miRNA lies in an intron of the gene qtc. The genome browser indicate that the simulans alignment in this area is ambiguous as evident from chains in the browser screenshot below. As it appears, the ortholog for qtc would lie between a rearrangement event with chrU, chr3R, and chr2L_random:
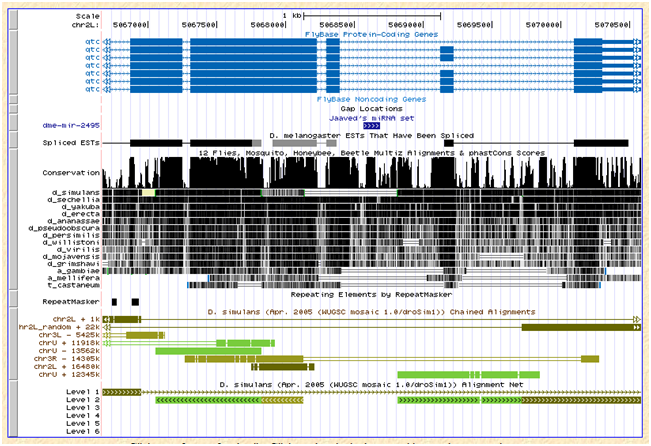
Flybase reports an ortholog of dsim\GD23320 for qtc, but this annotation was recently withdrawn. No ortholog is reported for sechelia but for erecta and yakuba. |
dme-mir-274 |
Dm3 |
Found |
N |
NA |
The correct ortholog was found in all the 7 strains. The correct simulans ortholog should lie in a gap region of the assembly (chr3L:11019633-11019769).
>dme-mir-274-dm3
AGTTTTGGTGACGAATCCTGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTGTTTGGCCGCCAGGATTACTCGTTTTTGCGATCACAAATTATGAAATTGCAGCAAAACTCAACGAAATTG
>zdo16e12.g1:275-405
AGTTTTAGTGACAAATCCTGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTGTTTGGCCACCAGGATTACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAATGCAACAAAAATG
|
dme-mir-280 |
Dm3 |
Found |
N |
NA |
The correct ortholog was found in all 7 strains. This ortholog should lie in a gap region of the simulans reference assembly (chr2R:2844465-2844618).
>dme-mir-280-dm3
CTGACGTGTGTATGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATATGCAAGTCGAGGTCCTCCACACTGCACTCGCCGCCTCGA
>zee54c10.b1:599-724
CTGACGTGTGTGTGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATATGCAAGTCGAGGTCCTGCACACTGCACTCGCCGCCTCGA |
dme-mir-281-2 |
Dm3 |
Found |
N |
NA |
The correct ortholog was fully found in 3 of the 7 strains, and partly found in the remaining 4. This ortholog should lie upstream to the dme-mir-281-1 ortholog, but this region is actually filled by a gap (chr2R_random:2147066-2148065). The MAF actually reports the 281-2 and 281-1 orthologs as the same, but this is clearly not the case apparent from the discordant substitution patterns.
>dme-mir-281-2-dm3
CTTTCAGATCCTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTAAGACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGATTAT
>zdp42d09.g1:294-416
CTTTCAGATCCTCCGCGAATTGTGAAATGAAGAGAGCTATCCGTCGACAGTCAAGTTAAGACCGATTGTAATACTGTCATGGAATTGCTCTCTTTGTATAACATTCGAAAGGCGACGATTAT |
dme-mir-306 |
Dme |
|
N |
NA |
The correct ortholog was found in its entirety in 6 of the 7 strains, and partially found in md106.
>dme-mir-306-dm3
GTGAATAGTTTAAAAGTCCACTCGATGGCTCAGGTACTTAGTGACTCTCAATGCTTTTGACATTTTGGGGGTCACTCTGTGCCTGTGCTGCCAGTGGGACATAATCTACAAATAA
>zdv96e12.b1:525-640
GTGAATAGTTTAAAAGTCCACTCGATGGCTCAGGTACTTAGTGACTCTCAATGCCTTTGACATTTTGGGGGTCACTCTGTGCCTGTGCTGCCAGTGGGACATAATCTACAAATAA
This miRNA lie in the same intron of the grp gene as 79 and 9b, and one intron downstream of 9c. The genome browser show that this region's alignment with simulans is ambiguous:
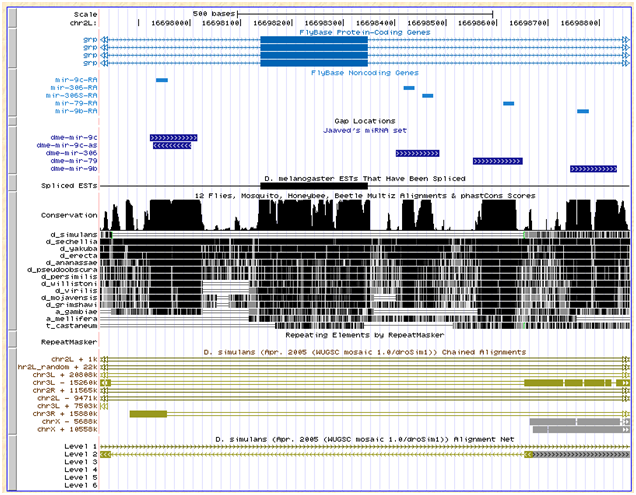
Flybase also does not report an ortholog for grp in simulans, however one exists for sechellia (GM17194). |
dme-mir-79 |
Dm3 |
Found |
N |
NA |
The correct ortholog was only found in 4 of the 7 species. It was found partially in 1 (c1674) and not found in 2 (sim4 & md106).
>dme-mir-79-dm3
ATGAGTGCCTTAGAGTGAAGCTGACTTGCCATTGCTTTGGCGCTTTAGCTGTATGATAGATTTAAACTACTTCATAAAGCTAGATTACCAAAGCATTGGCTTCTGCAGGTCAATCGTCAGAAACAAT
>wck39a12.g1:600-726
ATGAGTGCGTCAGAGTGTAGCTGACTTACCATTGCTTTGGCGCTTTAGCTGTATGATAGATTTAAACTACTTCATAAAGCTAGATTACCAAAGCATTGGCTTGTGCAGATCAATCGTCAGAAACAA- |
dme-mir-971 |
Dm3 |
Found |
N |
NA |
>dme-mir-971-dm3
TGCATGTGAGAGAATTCCGTGGCTGGCATCGCTCGCTGTAAATTGTAATCATCAAAGCGTTTTCTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCCGTACACAGAAAGAAAACC
>zec46h04.g1:44-170
TGCATGTGAGAGAATTCCGTGGCTGGCATCGCTCGCTGTAAATTGTAATCATCAAAGCGTTTTCTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCCGTACACAGAAAGAAAACC
This reads lie in a gap in the simulans assembly (chrX:9859862-9862861). The sequence was not found in the w501 strain which comprises the bulk of the droSim1 assembly. It was found in the 6 other strains. |
dme-mir-9b |
Dm3 |
Found |
N |
NA |
The best ortholog was found in the nc48 strain. Other suboptimal hits were found in c1674, md106, md199, and sim6. No hits found in sim4 or w501
>dme-mir-9b-dm3
TGTTGCTCTTTTGTTTGCATATTATTTGCTCTTTGGTGATTTTAGCTGTATGGTGTTTATGTATATTCCATAGAGCTTTATTACCAAAAACCAAATGGTTTCTGCATTATGTTTGAGTTGA
>vom21g07.g1:730-849
TGTTGCTCTTTTGTTTGCATATTATTTGCTCTTTGGTGATTTTAGCTGTATGGTGTATAAGT--ATTCCATAGAGCTTTATTACCAAAAACCAAATGGTTTCTGCATTATGCTTGAGTTGA
|
dme-mir-9c |
Dm3 |
Found |
N |
NA |
>dme-mir-9c-dm3
CTTGCACTATTTATCATTTTTGCTGTTTCTTTGGTATTCTAGCTGTAGATTGTTTCACGCACATTGTATATCATCTAAAGCTTTTATACCAAAGCTCCAGCTTAAATTGCTTAACATGATAT
>zdv96e12.b1:15-131
CTTGCACTATTTATCATTTTTGCTGTTTCTTTGGTATTCTAGCTGTAGATTGTTTCACGCACATTGTATATCATCTAAAGCTTTTATACCAAAGCTCCAGCTTAAATTGCTTAACA------
All strains contained the ortholog. |
dme-mir-4954 |
Dm3 |
Found |
N |
NA |
This miRNA lies in an intron of the dm3 gene actn. Actn has no reported ortholog in simulans. The Alignment in this area in simulans is also ambiguous; there are several chains in chrU, chr3L and chrX that comprise the surrounding syntenic region.
The ortholog was only found in w501 strain, partially in nc48, and none of the other 5 strains
>dme-mir-4954-dm3
ATATAAGTGTTCGATTTGGCGCTTGGAATCGATACCCGAGCCATGATAGATTGAAGTCAACCCAATCGATCGCGGTTCGAGTGCTCGAGTCTTGTGCGCCGGCATGCTTGAGATGTCCACTA
>vgb65a08_5.b1:263-385
ATATAAGTGTTCGATTTGGCGCTTGGAATCGATACCCGAGCCCTGATAGATTGAAGTCAACCCAATCGATCGCGGTTCGGGTGCTCGAGTCCTGTGCGCCGGCATGCTTGAGATGTCCACTA
|
dme-mir-4958 |
Dm3 |
Found |
N |
NA |
The ortholog was only found in md106 strain and in none of the other 6.
>dme-mir-4958-dm3
GTCCGAACCGTACGATCTCTGGCAGCTGCAGTTCCGTTTCCGGAGCGGGATCTGGAACGGGCTCTGGTTCTAGATCAGGATCCAATCATGCACCCGGTCCAGGTACCGCTCCAGGTCCCGTTCCCGGAAACGGTGCCACCGCCAATGCAGCTGCAGCGGCATTCA
>wgm54c09.g1:392-557
GTCCGAACCGCACGATCTCTGGCAGCTGCAGTTCCGTTTCCGCAGCGGGATCTGGGACGGGCTCTGCTTCTAGATCAGGATCCAATCATGCACCCGGTCCAGGTGCCGCTCCAGGTCCCGTACCCGGAAGCGGAGCCACCGCCAATGCAGCTGCAGCGGCATTCA |
dme-mir-4961 |
Dm3 |
Found |
N |
NA |
This miRNA is on the 3' UTR of the CG3056 gene. The ortholog for this gene is dsim\GD16553. This gene is very short (1KB), and had no similarity with 4961/dme.
On the browser, the syntenic region of this miRNA would lie in a gap (chrX:979887-981047). However, searching the reads, the ortholog is found:
>dme-mir-4961-dm3
AGATATTACCCAGAAGCGATATCCAATAGTAGCCAACTCTCTCGCTCTCTATGTGTATGTATGTATCTTGCTATCCATATATGTATATCCATATCAGAGAGCCAGGATTGGTTGAGACCATACGATATAACCCGAAACCACACTGGCCAGGAATAGCAAATCCA
>wjj76b12.b1:303-467
AGATATTACCCAGAAGCGATATCCAATAGTAGCCAACTCTCTCGCTCTCTATGTGAATGTATGTATCTTGCTATCCATATATGTATATCCATATCAGAGAGCCAGGATTGGTTGAGACCATACGATATAACCCGAAACCATACTGGCCAGGAATAGCAAATCCA
The best hits were found in all the strains except for w501 and c1674. In w510 only a partial hit was found.
|
dme-mir-4971 |
Dm3 |
Found |
N |
NA |
This miRNA actually lies in the CDS region of msl-2 gene. No ortholog in simulans is reported but an ortholog in sechelia exists.
>dme-mir-4971-dm3
GGAAGGGCCAACTTCTCGGCCCTCGACACGGTGGATGAGCTTGTCAGTGGCGGATCCAGGAGCAATTCTGCCGCTGGCGACAGATCATCGGCCACTGACAATGCCCATTCACTGTTCGAGGAGATCATGTCGGGCTCGGATG
>wop21h10.b1:281-423
GGAAGGGCCAATTTCTCGGCTCTCGACACGGTTGATGAGCTTGTCAGTGGCGGATCCAGGAGCAATTCTGCCGCTGGCGACAGATCATCGGCCACTGACAATGCCCAGTCACTGTTCGAGGAGATTATGTCGGGCTCGGATG |
miRNA Name |
Query Species |
Result |
Fragment to clone |
Experiment Needed |
Experiment Decision |
Sequence |
dme-mir-12 |
dm3 |
Found. |
|
N |
NA |
>dme-mir-12-dm3
AAGGAGCAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA
>gnl|ti|371645083:614-718
AAGGAGCAGCGTCTGTACGGTTGAGTATTACATCAGGTACTGGTGTGCCTTAAATCCAACAACCAGTACTTATGTCATACTACGCCGTGCACGGATCGCACTAA
|
dme-mir-972 |
dm3, droEre2 |
Not found |
chrX:11892021-11893235
(1214nt) |
Y |
Requenced - Found the 973 and 974, but not 972 |
chrX:11892021-11893235 has a 1214nt gap.

The multiple alignment suggest that all three yakuba miRNA ortholog should lie within this gap, however, my search of the reads did not turn up any hits.
I should note that the assembly shotgun reads extend about 17-694nt into this gap, so it is mostly covered. Given that the miRNA downstream of this gap (975-977) has yakuba orthologs, it's very suspicious have this 972-974 miRNA sub-cluster loss. |
dme-mir-973 |
droSec1, droEre2 |
Not found |
dme-mir-974 |
droSec1, droEre2 |
Not found |
dme-mir-983-1 |
droSec1, droEre2 |
Not found |
|
N |
Genuine loss event. |
983-1/dme and 983-2/dme should lie in intron 5 of gene CG3626/dme. The ortholog for this gene is GRE16342/dyak. Searched this gene by BLAST for the mature and star sequence for both sechelia and erecta orthologs but only found hits of length 7 and not 21-23nt.
Only singleton reads (6 in total) from the yakuba small RNA head and female-body libraries reads mapped to the sechellia or erecta ortholog.
This looks like a genuine loss event. |
dme-mir-983-2 |
droSec1, droEre2 |
Not found |
|
N |
dme-mir-997 |
droSec1, droEre2 |
Not found |
Approximate genomic coordinates unknown. Clone with primers designed with dme-mir-997/dsec ortholog. |
Y |
Nothing done |
A search of the droYak2 genome assembly with the droSec1 ortholog found 15 hits, but these hits only captured the star sequence partially and did not span the mature sequence. droEre2 ortholog search within the droYak2 genome assembly did not return any hits.
>dme-mir-997-droSec1
CAATAAATATGTAGTTTTAGATACTCGCCAGTCAGGATGCTCTGTCAATGAATTTAGTATGCCCAAACTCGAAGGAGTTTCACCTCCATAGGAGCGGCAGACCTGGAGAAGTTTTCAGAGCCAACAAAATTCATATGATGATGCATTTTCAGTCTCTGAAAATTTCTTCAGCAGAAGTTGATTTTAGCGAAGTGAAGCTCATTCGATTTTGATCATACTAACATTTGTGGATGCTTGGATCGTCAGTTTGGTGGAATATT
>gnl|ti|386447040:428-513
CG-------------------------------------------------------------------------------------------------------------------------ACACAATTTATTTAATGATGCATTTTCAGTCCCTGAAATCTTCTTCAGTAAAAGTGGATTTTTGTAAAGTTGAGCTCATTCGA------------------------------------------------------
The erecta ortholog has many substitutions that don't agree closely with the simulans nor sechellia ortholog, however this ortholog folds into a long hairpin and has reads within the mature and star regions indicative of RNase III cleavage.
Do the yakuba reads map to the sechellia ortholog?
Yes. 14 yakuba small RNA reads mapped to the sechellia ortholog, however all were 19nt long. Since the orthologs reported in the MAF and by LASTZ searches do not appear confident and the hit reported from trace reads search return a gappy, non-confident sequence, we are unsure what the correct region to PCR would be. This ortholog appears to be lost.
V052_mapped_droSec1.sam:V052_199748_1 16 dme-mir-997_droSec1 65 25 19M * 0 0 AACCTCGAAGCAGTTTCAC*XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2A7G8
V052_mapped_droSec1.sam:V052_455157_1 16 dme-mir-997_droSec1 43 25 19M * 0 0 TGTCAATGAATGCAGTATG*XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:11T0T6
V058_mapped_droSec1.sam:V058_51901_5 16 dme-mir-997_droSec1 65 25 19M * 0 0 AAACTCTAAGCAGTTTCAC*XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:6G3G8
V058_mapped_droSec1.sam:V058_161246_2 16 dme-mir-997_droSec1 65 25 19M * 0 0 AACCTCGAAGCAGTTTCAC*XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2A7G8
V058_mapped_droSec1.sam:V058_169511_2 16 dme-mir-997_droSec1 66 25 18M * 0 0 AACTCTAAGCAGTTTCAC *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5G3G8
V058_mapped_droSec1.sam:V058_701167_1 16 dme-mir-997_droSec1 129 25 18M * 0 0 ATTCAGATGGTGATGCAT *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T3A8
V058_mapped_droSec1.sam:V058_891376_1 16 dme-mir-997_droSec1 83 37 5M1I12M * 0 0 CATCCGATAGGAGCGGCA *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:1C15
V058_mapped_droSec1.sam:V058_1231934_1 16 dme-mir-997_droSec1 66 25 18M * 0 0 ACCTCGAAGCAGTTTCAC *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:1A7G8
V058_mapped_droSec1.sam:V058_1365009_1 16 dme-mir-997_droSec1 66 25 18M * 0 0 AGCTCGAAGCAGTTTCAC *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:1A7G8
V058_mapped_droSec1.sam:V058_1866832_1 16 dme-mir-997_droSec1 65 25 19M * 0 0 AAACTCAAAGCAGTTTCAC*XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:6G3G8
M026_mapped_droSec1.sam:M026_181245_1 16 dme-mir-997_droSec1 197 25 18M * 0 0 GCTCAATCGACTTTGATC *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T4T7
M026_mapped_droSec1.sam:M026_330923_1 16 dme-mir-997_droSec1 129 25 18M * 0 0 ATTCAGATGGTGATGCAT *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T3A8
M043_mapped_droSec1.sam:M043_137968_23 16 dme-mir-997_droSec1 129 25 18M * 0 0 ATTCAGATGGTGATGCAT *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T3A8
M043_mapped_droSec1.sam:M043_707351_3 0 dme-mir-997_droSec1 135 25 19M * 0 0 ATGATGATGATTTTTCAGT*XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:9C0A8
M043_mapped_droSec1.sam:M043_2823216_1 0 dme-mir-997_droSec1 238 25 18M * 0 0 GATCGTCAGTGTGCTGGA *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:10T2G4
M043_mapped_droSec1.sam:M043_3233721_1 16 dme-mir-997_droSec1 13 25 21M * 0 0 AGCTTTAGATACTCGCGAGTC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2T13C4
M043_mapped_droSec1.sam:M043_5112769_1 0 dme-mir-997_droSec1 223 37 9M1I8M * 0 0 ATCTGTGGATTGCTTGGA *XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:2T14
V120_mapped_droSec1.sam:V120_98271_7 0 dme-mir-997_droSec1 135 25 19M * 0 0 ATGATGATGATTTTTCAGT * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:9C0A8
V120_mapped_droSec1.sam:V120_961113_1 16 dme-mir-997_droSec1 141 25 19M * 0 0 ATGCATTTTCAGTCTTTGC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:15C2A0
V120_mapped_droSec1.sam:V120_1656829_1 0 dme-mir-997_droSec1 116 37 12M1I5M * 0 0 CAGAGCCAACAACAATTA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:16C0
|
miRNA Name |
Query Species |
Result |
Fragment to clone |
Experiment Needed |
Experiment Decision |
Sequence/Notes |
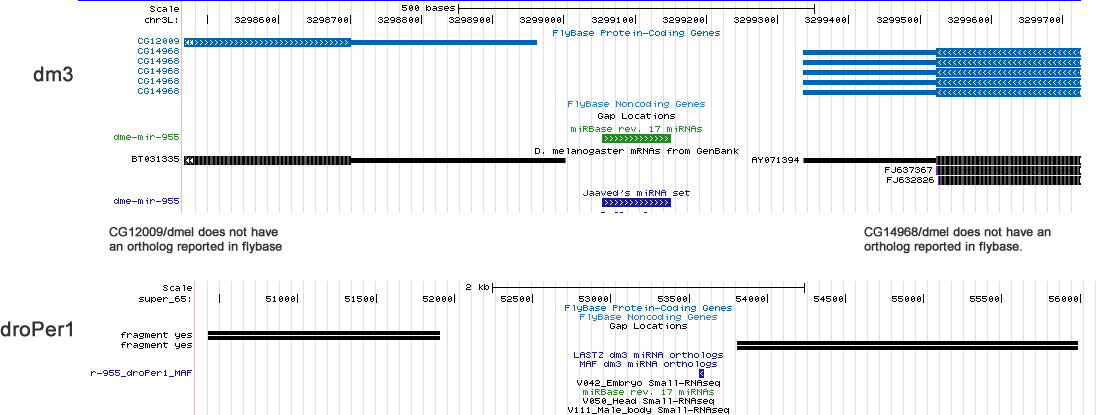
dme-mir-313 |
droAna3 |
Not found |
chr3:8678956-8680852
(1897nt)
This region should emcompass dme-mir-992/dpse - dps-mir-2507b |
Y |
Cluster will be resequenced |
The alignment suggest that 313/dpse should lie adjacent to 312/dpse, similar to how 313/dper lies adjacent to 312/dper. However, searching the dp4 assembly with the annanasae ortholog did not turn up any hits. In pseudobscura, the 310-313 cluster has many adjacent miRBase annotated miRNAs. With so many new annotations, this locus may contain many duplicate miRNAs and thus poorly assembled. Therefore, resequencing is needed to clarify if 313/dpse is genuinely missing.

|
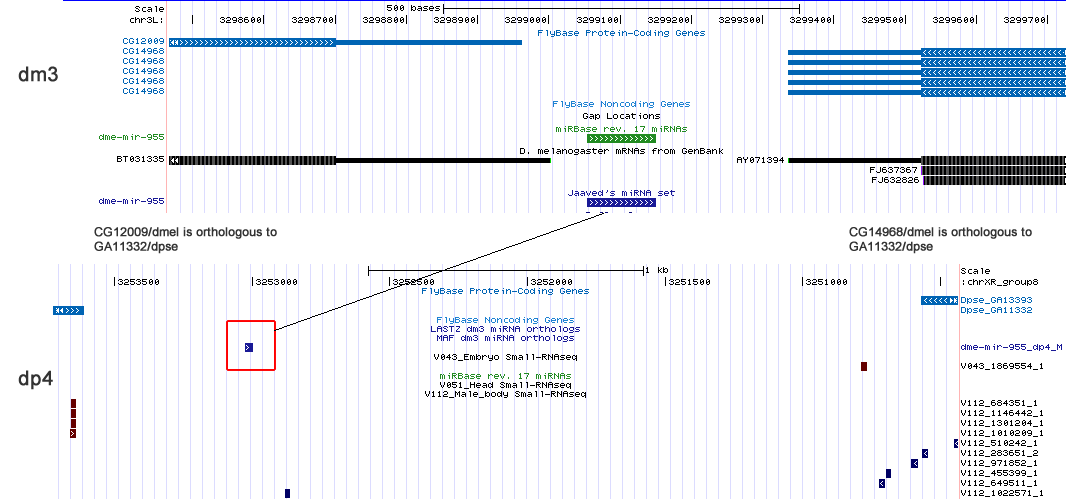
| dme-mir-955 |
droAna3 |
N |
chrXR_group8:3250437-3253723 -
(3286nt) |
N |
Confidently lost in the obscura subgroup |
955/dmel is an intergenic miRNA. The flanking genes in melanogaster have orthologs in pseudobscura as well. Similarly, the 955/pse ortholog is also intergenic and flanked by these orthologous genes.
A search of the pseudoobscura sRNAseq libraries mapped to the ananassae and willistoni ortholog turned up few reads. Are are the reads mapping to the willitoni ortholog:
V043_mapped_droWil1.sam:V043_73838_4 16 dme-mir-955_droWil1 62 37 6M1I11M * 0 0 TTTGTTGTTCTCCAATGG
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:11T5
V043_mapped_droWil1.sam:V043_149175_2 0 dme-mir-955_droWil1 35 25 22M * 0 0 CATCGTGCACAGGTTTGAGTGT * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:8T0T12
V043_mapped_droWil1.sam:V043_169017_2 4 dme-mir-955_droWil1 116 25 20M * 0 0 AACAGCCTAAAAAGTACAAT * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:12T2G4
V043_mapped_droWil1.sam:V043_1068829_1 0 dme-mir-955_droWil1 16 25 18M * 0 0 CATTAAGGATGGCTGGCT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:1G6T9
V043_mapped_droWil1.sam:V043_1265617_1 0 dme-mir-955_droWil1 35 25 23M * 0 0 CATCGTGCAGAGGTTTGAGTGTC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:8T0T13
M040_mapped_droWil1.sam:M040_2435810_2 0 dme-mir-955_droWil1 61 25 18M * 0 0 TTTTGTTTTCGCTAATGG
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:0G9T7
M040_mapped_droWil1.sam:M040_3794921_1 0 dme-mir-955_droWil1 16 25 18M * 0 0 CATTAAGGATGGCTGGCT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:1G6T9
M040_mapped_droWil1.sam:M040_3943027_1 16 dme-mir-955_droWil1 49 37 8M1D11M * 0 0 TTGAGTGTTTCGTTCGTTT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:8^C6T4
M040_mapped_droWil1.sam:M040_4187516_1 0 dme-mir-955_droWil1 51 25 18M * 0 0 GAGTGACTTCCTTTGTTT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T4G7
M040_mapped_droWil1.sam:M040_4279699_1 4 dme-mir-955_droWil1 117 25 19M * 0 0 ACAGCCTAAAAAGTACAAT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:11T2G4
M040_mapped_droWil1.sam:M040_4468108_1 0 dme-mir-955_droWil1 62 37 5M1I13M * 0 0 TTTGTGTTTCTCAAATGGC
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:11T6
M040_mapped_droWil1.sam:M040_5795261_1 0 dme-mir-955_droWil1 43 37 12M1I5M * 0 0 TTAGGATTGAGTCGTCTT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:5T11
M040_mapped_droWil1.sam:M040_9979667_1 0 dme-mir-955_droWil1 24 37 9M2I9M * 0 0 TTGGCTGGCCGTCCATCGTG * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:0 XO:i:1 XG:i:2 MD:Z:18
This miRNA looks confidently lost in obscura lineage.
|
dme-mir-963 |
droAna3, droWil1 |
Not found |
|
N |
Confidently lost |
See notes in persimilis.
I checked how many pseudoobscura reads mapped to the willistoni and ananassae orthologs, but found very few did and they did not map perfectly. Here are the reads mapping to the willistoni ortholog:
V112_mapped_droWil1.sam:V112_335143_2 16 dme-mir-963_droWil1 123 25 20M * 0 0 TTTTCGTTAAGCCCTACACA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T8A5
V112_mapped_droWil1.sam:V112_349332_2 0 dme-mir-963_droWil1 95 25 21M * 0 0 TAGCTTTGTTTCGTATAGGAC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2C10A7
V112_mapped_droWil1.sam:V112_941426_1 16 dme-mir-963_droWil1 123 25 21M * 0 0 TTTTCGTTAAGCCCAACACAA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5T14T0
V112_mapped_droWil1.sam:V112_1298991_1 16 dme-mir-963_droWil1 122 25 21M * 0 0 TTTTTCGTTAAGCCCAACACA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:0G5T14
V043_mapped_droWil1.sam:V043_198574_1 0 dme-mir-963_droWil1 27 37 10M1I7M * 0 0 TAATTCTAGTGCTAATAC
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:14A2
V051_mapped_droWil1.sam:V051_563156_1 16 dme-mir-963_droWil1 73 25 19M * 0 0 TGGAGCCTGAAACATCCGT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2A13T2
M040_mapped_droWil1.sam:M040_4092641_1 16 dme-mir-963_droWil1 22 37 9M1I10M * 0 0 AATTCTAATCTCTCGTCTAA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:12A6
M040_mapped_droWil1.sam:M040_4593573_1 0 dme-mir-963_droWil1 68 37 10M1I7M * 0 0 ATATTTGGAGACCTGAAA
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:7A9
M040_mapped_droWil1.sam:M040_6556843_1 0 dme-mir-963_droWil1 42 37 21M * 0 0 ACAAGGTACATATCAGGTTGT * XT:A:U NM:i:0 X0:i:1 X1:i:0 XM:i:0 XO:i:0 XG:i:0 MD:Z:21
M040_mapped_droWil1.sam:M040_6571014_1 20 dme-mir-963_droWil1 143 37 12M1D7M * 0 0 TTTGTTTTTACGCCAACAC
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:10A1^C7
M040_mapped_droWil1.sam:M040_7361873_1 20 dme-mir-963_droWil1 147 25 18M * 0 0 TTTTTAAGGCGAACACAT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:8C1C7
M040_mapped_droWil1.sam:M040_9413604_1 0 dme-mir-963_droWil1 84 25 22M * 0 0 ACATCTGTATATACCTTTGTTC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:10C10T0
This miRNA appears confidently lost in the obscura lineage. |
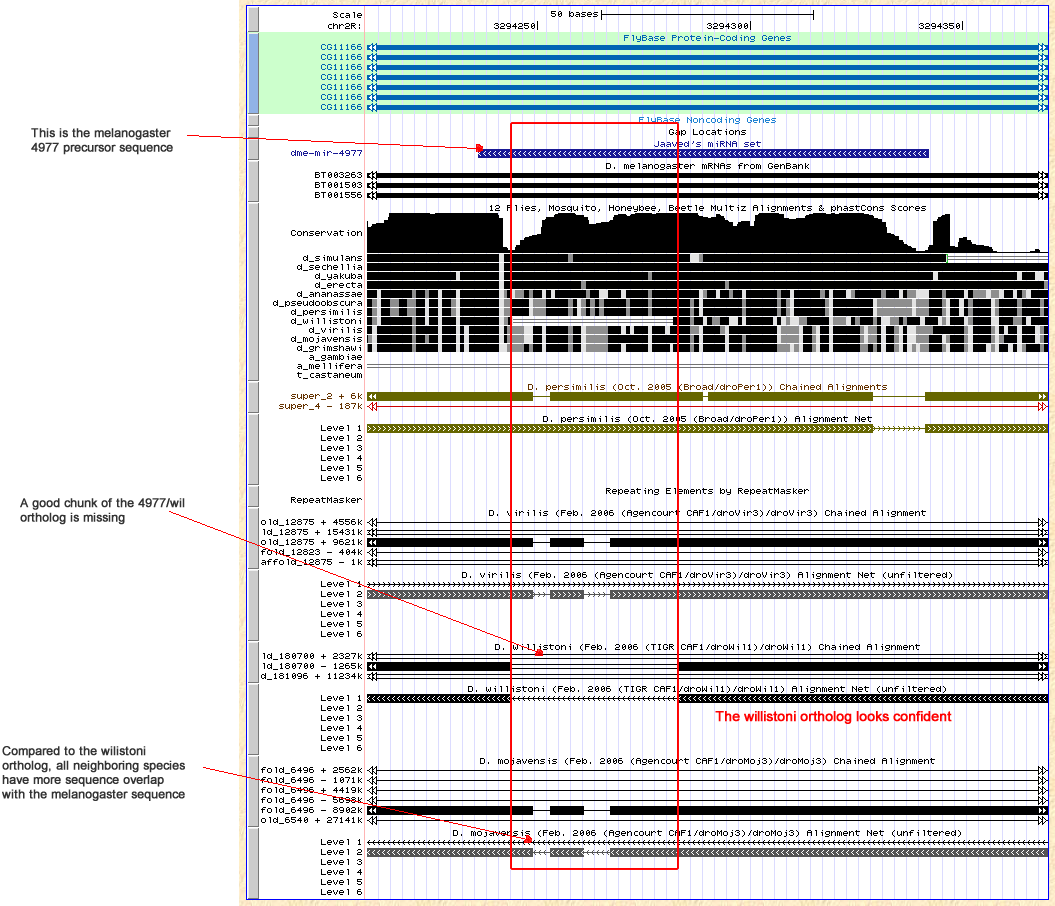
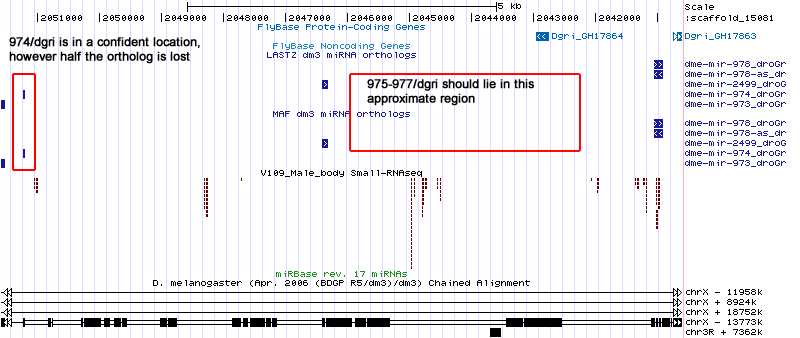
| dme-mir-973 - 978 |
-- |
Not found |
Given that both flanking coding genes are on different chromsomes, we may be able to define forward and reverse primers on different chrsomosomes, however it is still unsure how large the intermediate region will be. |
Y |
Nothing done |
These miRNAs lie in the intergenic region between CG32532/dmel and Grip84/dmel. CG32532/dmel has the ortholog GA23014/dpse (chrXL_group1a:5357217-5385309) and Grip84/dmel has the ortholog GA17771/dpse (chrXL_group1e:12505277-12510040 +). These two orthlogous genes lie on different chromsomes. In fact, GA17771/dpse lies 15Kb from the end of chrXL_group1e. It is highly suspicioius that these miRNA orthologs are missing given the choppy assembly in this region. |
miRNA Name |
Query Species |
Result |
Fragment to clone |
Experiment Needed |
Experiment Decision |
Sequence/Notes |
dme-mir-2500 |
dp4 |
Found |
|
N |
NA |
Dm3 miRNA lies within CG3764/dme. This gene has an ortholog reported in dp4, namely dpse\GA17671, but none reported for droPer1 according to Flybase.
According to the nets and chains track for droPer1, the ortholog should lie in a stretch of ambiguity characters (N's) at position super_29:1057504-1057925. Searching the droPer1 trace reads with the dp4 ortholog correctly locates the ortholog.
>dme-mir-2500-dp4
AAGAACTTTTTGCAGGTAAGCCATCACCAGCATTGAAAGCCATTGAAATGCGTCTAAATAGCAGTCTCTCTCCTCCCACAGATCATGTTCCAGGCC
>gnl|ti|780630171:515-611
AAGAACTTTTTGCAGGTAAGCCATCACCAGCATTGAAAGCCATTGAAATGCGTCTAAATAGCAGTCTCTCTCCTCCCACAGATCATGTTCCAGGCC |
dme-mir-285 |
dp4 |
Found |
|
N |
NA |
This miRNA is intergenic and there appear to be no nearby genes. The correct persimilis ortholog is found by searching the persimilis trace reads for the pseudobscura sequence via LASTZ.
>dme-mir-285-dp4
TCTTACAAT-TACAGTTCGAATCGAAGAAC-TGAGATCGATTGGTGCATAGATATCAAGAGGACTCGCTAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA
>gnl|ti|738578230:636-763
TCTTACAATTTACAGTGGGAATCGAAGAACCTGAGATCGATTGGTGCATAGATATCAAGAGGACTCGATAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA |
dme-mir-304 |
Dp4, droWil1 |
Not found |
super_26:1090127-1093182
(3056nt) |
Y |
1st round of resequencing indicates that the ortholog may lie immediately before the gap, but the quality of the returned sequence was poor. |
A search of the persimilis trace data with the dp4 or droWil1 orthologs did not return any hits. This miRNA lies in an intron of Gmap/dmel. It is unusual that flybase did not report an ortholog for this gene in persimilis but did for pseudobscura (GA22886/dpse). 304 lies on the same intron as 283/dme and 12/dme and the droPer1 ortholog for these were found; this is the first peice of evidence to suggest that the ortholog exists.
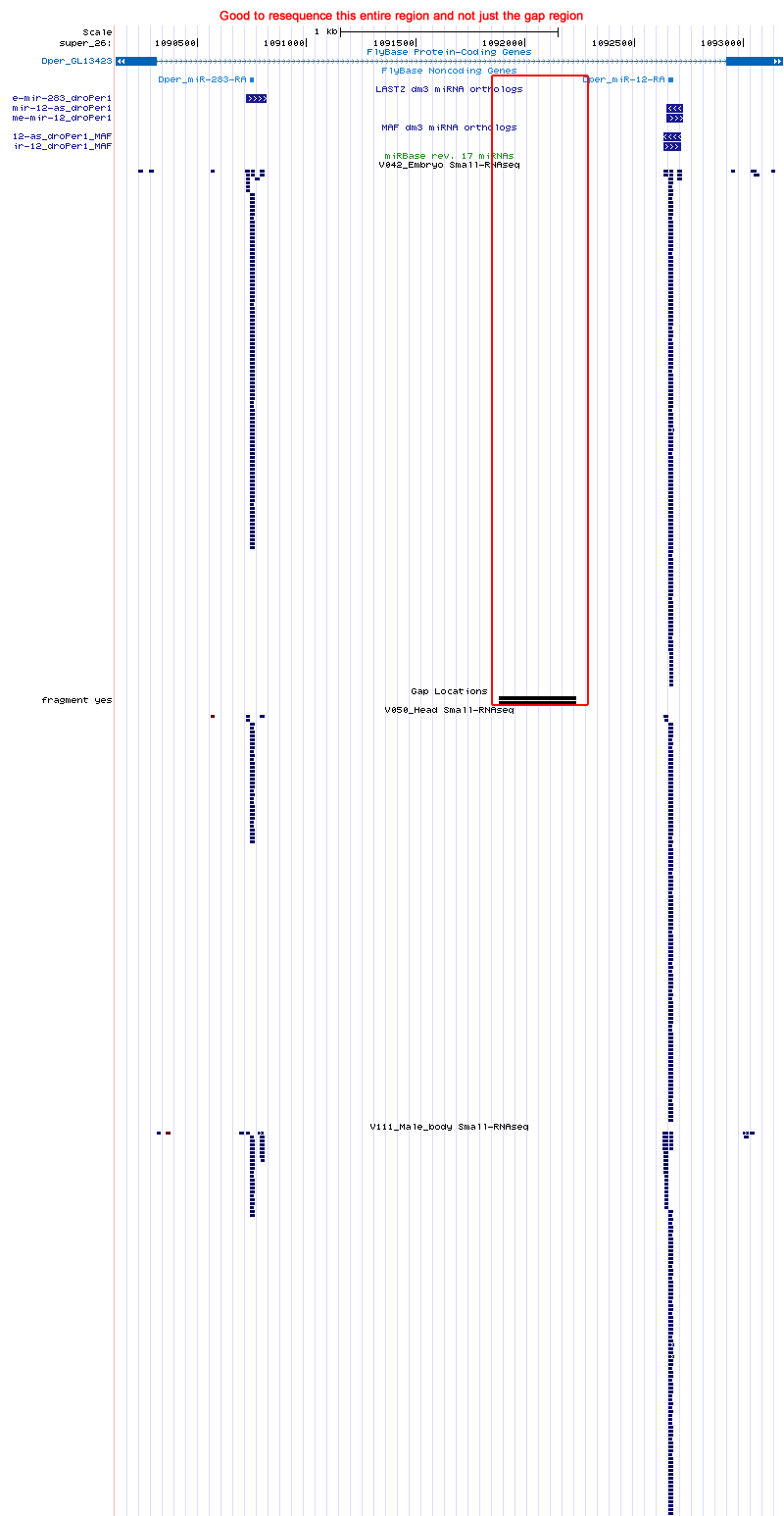
The flanking synthenic regions of dp4's ortholog offer some clues as to where the droPer1 ortholog may lie. droPer1 ortholog may lie in a gap region located at super_26:1091883-1092236. This gap is 353nt long. The two neighboring assembly read only go 77nt and 28nt into the gap from either side, but it seem like the miRNA lies more toward the middle of this gap and thus uncovered by reads.
Does the persimillis reads map to the pseudobscura lineage?
Yes. Reads from all persimilis libraries align to the dp4 ortholog. For example V042_1404_168, V111_2403_579, and V050_394_390. These reads are all 23nt in length and contain the DNA sequence "TAATCTCAATTTGTAAATGTGAG". These reads are unmapped to the droPer1 genome assembly genome assembly. The DNA sequence of these reads also do not mapp to the trace reads for droPer1. |
dme-mir-311 |
Dp4 |
Found |
|
N |
Cluster will be resequenced anyways |
>dme-mir-311-dp4
CATCGAGCTGCTTTGATTTGTAGGCCGTGGTTCTTGCAAATACGGATTCATAACGTATTGCACTAGCCCCGGTCCAAAAAACAATAGCAACGCCGGCAACAGCAAA
>gnl|ti|732560583:591-697
CATCGAGCTGCTTTGATTTGTAGGCCGTGGTTCTTGCAAATACGAATTCATAACGTATTGCACTAGCCCCGGTCCAAAAAACAATAGCAACGCCGGCAACAGCAAA
When this droPer1 sequence is blatted against the droPer1 assembly, reads pileup nicely on both mature and star regions (super_4:2,566,608-2,566,713). The MAF does not report this region as the ortholog to the dm3 sequence. Instead, the MAF reports that this sequence partly belongs to 312/dper. 312/dper looks genuinely missing and is confident in pseudobscura, willistoni, and grimshawi. |
dme-mir-312 |
Dp4 |
Found |
|
N |
Cluster will be resequenced anyways |
312/dper looks genuinely missing but is confident in pseudobscura, willistoni, and grimshawi. Searching with the 312/dpse actually finds the correct persimilis sequence which also turns out to be 313/dpse.
>dme-mir-312-dp4
GGGCAATACTGTGTTGTATTTCACCAGTATTGCACACCCACTGGCCTGAAAGTGCCTACTGCTGGGTTCAAA
>gnl|ti|732455623:23-94_313_dpse
GGGCAATACTGTGTTGTATTTGAACAGTATTGCACACCCACTGGCCTGAAAGTGCCTACTGCTGGGTTCAA-
|
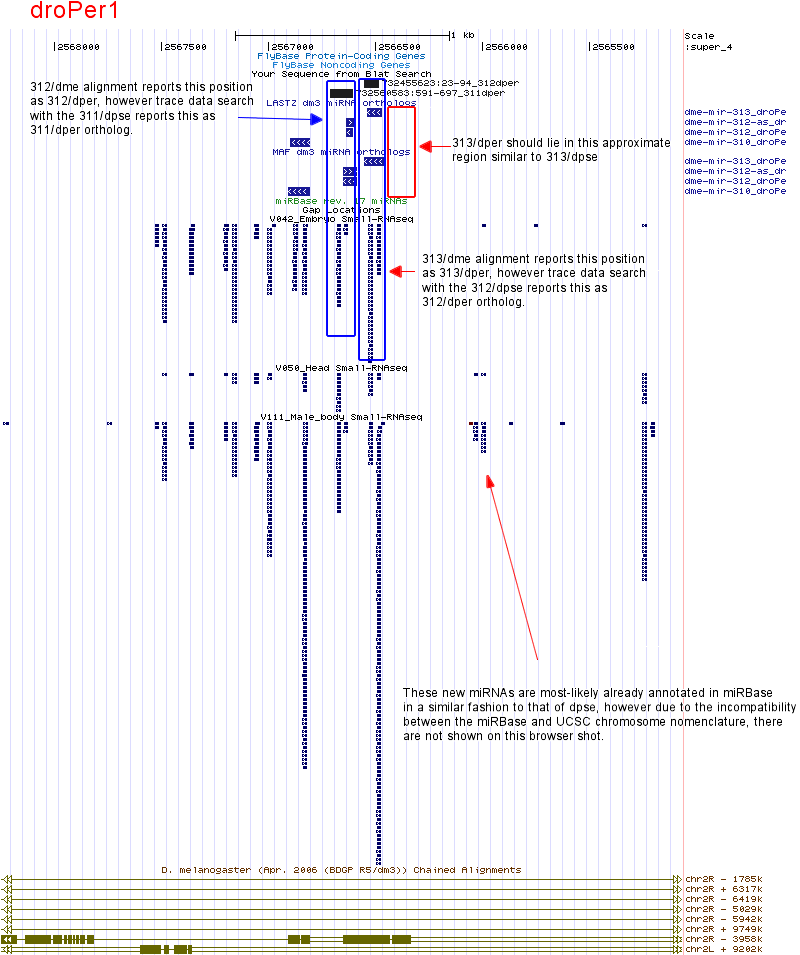
| dme-mir-313 |
droAna3 |
Not found |
super_4:2565122-2567706
(2584nt) |
Y |
Cluster will be resequenced |
Similar to 313/dpse, this ortholog is also missing. Searching the droPer1 assembly with the annanasae ortholog also did not turn up any hits. Given that species that outgroup the obscura lineage contain orthologs for this miRNA, resequencing is need to validate if the 313/dper ortholog is genuinely lost. It also would not hurt to resequence this enter cluster within both obscura species.
|
dme-mir-955 |
droAna3, droWil1 |
Not found |
super_65:50349-56133
(5784nt) |
N |
Confidently lost in obscura subgroup. |
955/dmel lies in an intergenic region. The flanking coding genes to this miRNA in melanogaster has no reported ortholog in persimilis. However, it is unusual to have orthologs reported in pseudobscura and not in persimilis. Looking at the persimilis assembly suggest that the homologous flanking genes may lie in the gap regions below..
Searching the trace data with droAna3 or droWil1 orthologs turned up very limited number of reads. Here are the reads mapping to the willistoni ortholog:
V111_mapped_droWil1.sam:V111_801966_1 0 dme-mir-955_droWil1 35 25 23M * 0 0 CATCGTGCAGAGGTTTGAGTGTC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:8T0T13
V111_mapped_droWil1.sam:V111_974686_1 0 dme-mir-955_droWil1 35 25 19M * 0 0 CATCGTGCAGAGGTTTGAG
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:8T0T9
V042_mapped_droWil1.sam:V042_610002_1 4 dme-mir-955_droWil1 118 25 18M * 0 0 CAGCCTAAAAAGTACAAT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:10T2G4
M021_mapped_droWil1.sam:M021_65469_10 16 dme-mir-955_droWil1 62 37 6M1I11M * 0 0 TTTGTTGTTCTCCAATGG
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:11T5
M021_mapped_droWil1.sam:M021_104734_6 0 dme-mir-955_droWil1 16 25 18M * 0 0 CATTAAGGATGGCTGGCT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:1G6T9
M021_mapped_droWil1.sam:M021_1027206_1 0 dme-mir-955_droWil1 10 37 12M1D7M * 0 0 CTTTTGCGTTAAGTTGACT
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:12^G4G2
M042_mapped_droWil1.sam:M042_1599213_1 16 dme-mir-955_droWil1 99 37 5M1D15M * 0 0 GAGACGCAAATCGAGAAACA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:5^G2C12
M042_mapped_droWil1.sam:M042_1665682_1 0 dme-mir-955_droWil1 49 37 3M1I14M * 0 0 TTGAAGTGTCCTCGTTTG
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:9T7
This miRNA looks confidently lost in the obscura subgroup. |
dme-mir-956 |
Dp4 |
Found |
|
N |
NA |
>dme-mir-956-dp4
ATGTGCACTTCTACTGATCGTTATCGTGTTTGGAATGGTCTCGTTAGCTAACGGATGAGCAACTGCTTGCGCGCATTGGCCAAATGCATTTCGCACCGGGATGGATGTGGATGTGGCTGGATGTGGTGTCCCCAAGCACAGCCAGGTGTAGTTTCGAGACCACTCTAATCCATTGCACCGCCCACCCATTTATGCAAA
>gnl|ti|732454354:721-914
ATGTGCACTTCTACTGATCGTTATCGTGTTTGGAATGGTCTCGTTAGCTAACGGATGAGCAACTGCTTGCGCGCATTGG-CAAATGCATTTCGCACCG-GATGGATGTGGATGTGGCTGGATGTG-TGTTCCCAAGCACAGCCA-GTGTAGTTTCGAGACCACTCTA-TCCATTGCACCGCCCACCCATTTATGCAAA
|
dme-mir-963 |
droAna3, droWil1 |
Not found |
|
N |
Confidently lost in obscura lineage. |
This miRNA lie within the gene CG31646/dme. Flybase does not report an ortholog for this gene in the obscura lineage. The adjacent 964 has somewhat questionable orthologs, but it does have good read pileup on the mature and star strands.
Searching with the ananassae and willistoni orthologs in persimilis assembly reads returned very few reads which did not map perfectly. Here are the reads mapping to the willistoni ortholog:
V111_mapped_droWil1.sam:V111_95073_7 0 dme-mir-963_droWil1 42 25 24M * 0 0 ACAAGGTACATATCAGAGTGTTTC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:16G0T6
V111_mapped_droWil1.sam:V111_171720_3 0 dme-mir-963_droWil1 41 25 24M * 0 0 AACAAGGTACATATCAGAGTGTTT * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:17G0T5
V111_mapped_droWil1.sam:V111_799750_1 0 dme-mir-963_droWil1 42 25 23M * 0 0 ACAAGGTACATATCAGAGTGTTT * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:16G0T5
V111_mapped_droWil1.sam:V111_1107330_1 16 dme-mir-963_droWil1 23 25 20M * 0 0 ATCCTAATTCTCGTCTAAAA * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2T8A8
V111_mapped_droWil1.sam:V111_1107436_1 0 dme-mir-963_droWil1 42 37 24M * 0 0 ACAAGGTAAATATCAGGTTGTTTC * XT:A:U NM:i:1 X0:i:1 X1:i:0 XM:i:1 XO:i:0 XG:i:0 MD:Z:8C15
V042_mapped_droWil1.sam:V042_243826_1 20 dme-mir-963_droWil1 147 23 7M1I10M * 0 0 TTTTTAACGCCCAACACA
* XT:A:U NM:i:1 X0:i:1 X1:i:1 XM:i:0 XO:i:1 XG:i:1 MD:Z:17 XA:Z:dme-mir-963_droWil1,-126,7M1I10M,2;
V057_mapped_droWil1.sam:V057_1010865_1 16 dme-mir-963_droWil1 75 25 18M * 0 0 AAACCTCAAACATCTGTA
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:2G3G11
V057_mapped_droWil1.sam:V057_1016836_1 16 dme-mir-963_droWil1 75 25 18M * 0 0 AAGCCTGAACCATATGTA
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:9A3C4
M021_mapped_droWil1.sam:M021_406362_2 0 dme-mir-963_droWil1 60 37 4M1I13M * 0 0 TGTTGTCGTTTATTTGAA
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:8A8
M021_mapped_droWil1.sam:M021_715756_1 0 dme-mir-963_droWil1 92 37 4M1I15M * 0 0 ATCTGATCTTTGTTTCGAAT * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:5C13
M042_mapped_droWil1.sam:M042_1047908_1 0 dme-mir-963_droWil1 79 37 12M1I7M * 0 0 CTGGAACATCTGCTATCTAC * XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:1 XO:i:1 XG:i:1 MD:Z:3A15
M042_mapped_droWil1.sam:M042_3478115_1 0 dme-mir-963_droWil1 117 25 19M * 0 0 AAAGAAATTTCTTTAAGCC
* XT:A:U NM:i:2 X0:i:1 X1:i:0 XM:i:2 XO:i:0 XG:i:0 MD:Z:5G0T12
This miRNA appears confidently lost in the obscura lineage. |
dme-mir-973 - 978 |
droAna3, droWil1 |
Not found |
super_14:1988476-2012995
(24519nt)
Will have to PCR out this region in several fragments.
|
Y |
Nothing done |
These miRNAs lie in the intergenic region between CG32532/dmel and Grip84/dmel. CG32532/dmel has the ortholog GL14778/dper (super_14:2010114-2033134) and Grip84/dmel has the ortholog GL14777/dper (scaffold_14:1985718-1990982). Unlike the pseudobscura orthologs which lie on separate chromosomes, both of these orthologous coding genes lie on the same scaffold. However, there is no reads within this ~24kb region indicative of RNase III cleavage:
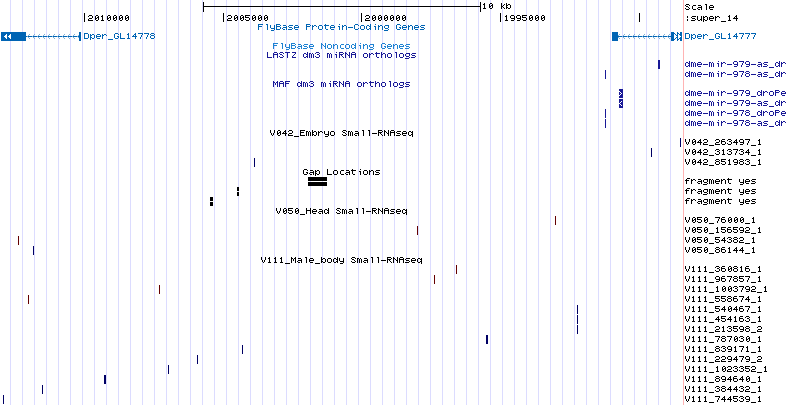
It is still uncertain whether these miRNAs are confidently lost in the obscura lineage. Some dp4 and droPer1 adult male-body reads do map to the droAna3 and droWil1 orthologs for all of these miRNAs suggesting that the correct obscura orthologs remain to be found. |
dme-mir-4984 |
Dp4 |
Found |
|
N |
NA |
>dme-mir-4984-dp4
TTAAAATAGTTGCAGCCCACAA-------GTCACATGCCGCCGCCGGCGCATTGGACATTGGAGGTGTGAAAACCAATATTCGCTTCAGGTCCAATCACGTTTCAAGATCGAGGTGAATTCTTTGACGTATTCGCTTGGCAAATGACACGTTTTGGACCTGCAATTTCCAGCATTTACTTTCATT
>gnl|ti|733036400:69-254
TTAAAATAGTTGCAGCCCACAAGTCACAAGTCACATGCCGCCGCCGGCGCATTGGACATTGGAGGTGTGAAAACCAATATTCGCTTCAGGTCCAATCACGTTTCAAGATCGAGGTGAATTCTTTGACGTATTCGCTTGGCAAATGACACGTTTTGGACCTGCAATTTCCAGCATTTACTTTCATT
Read gnl|ti|733036400 overlap with the adjacent gap at position super_1:4362270-4362410 |
miRNA Name |
Query Species |
Result |
Fragment for cloning |
Experiment Needed |
Experiment decision |
Sequence/Notes |
| dme-mir-312 |
droGri2 |
Found |
scaffold_12875:13668426-13669695
(1269nt) |
Y |
Cluster will be sequenced |
Searching with the grimshawi ortholog against the virilis genome trace reads found the sequence that is supposed to be 313/dvir.
>dme-mir-312-droGri2
AAGCAAATCTTCAGTTTTTCGGCTGTGGTTAGTGTCAATTCTTTTTAT-----TTGACAAGCATTGCACTCTTCACGGCCGGATAAAT-GAGGGTCT
>gnl|ti|391667200:98-186_313_droVir3
A------TCGCTAGTTTTTCGGCTGTGGTCAGTGTCAATTCTTTTTTTTTATTTTTACAAGTATTGCACTTTTCACGGCCGGAAAAATTGAGA-T--
This observation is actally also true for persimilis where the 313/dper ortholog was found when searching for the 312/dper ortholog with 312/dp4 query. |
| dme-mir-959 |
droWil1 |
Not Found |
scaffold_12963:3614884-3616293
(1409nt) |
Y |
Confidently lost |
Searching for the correct droVir3 ortholog with the droWil1 ortholog did not turn up any hits. It appears that 959, 960, and 961 are all lost in D. virilis.
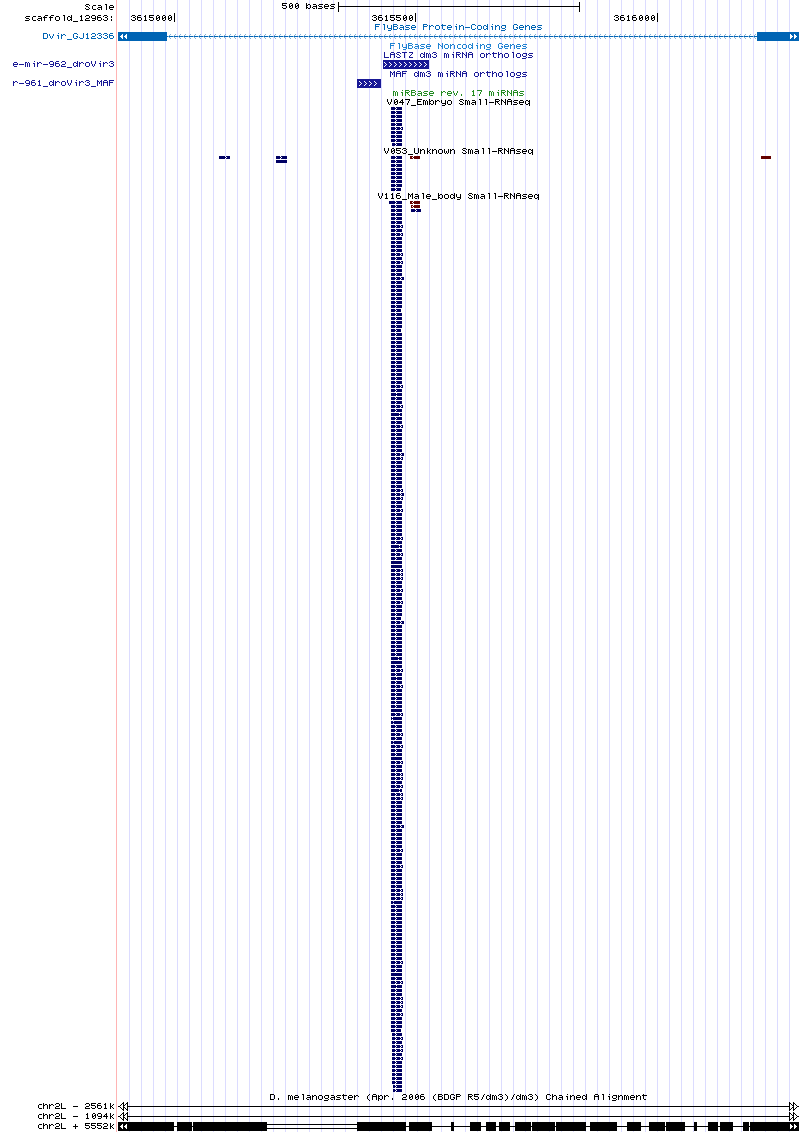
For 959 at least, the reported mojavensis and grimshawi orthologs are confident although neither of these orthologs has any small RNA reads
4 virilis reads mapped onto the 959/dwil ortholog. It would still be good to validate that this miRNA is lost in the virilis group. |
| dme-mir-960 |
droMoj3, droWil1 |
Not found |
Y |
Confidently lost |
Searching for the droWil1 or droMoj3 ortholog in droVir3 trace reads did not turn up any hits.
Do the virillis reads map to to the 960/dmoj ortholog?
There were only 5 droVir3 sRNAseq reads accross 3 libraries (Embryo, Male body and [Head?]) that aligned to the droWil1 ortholog. This suggest that this ortholog may be genuinely loss, but it would still be good to PCR out this entire cluster region in droVir3 and resequence it. |
| dme-mir-961 |
droWil1 |
Not Found |
Y |
Confidently lost. |
Searching for the correct droVir3 ortholog with the droWil1 ortholog did not turn up any hits. It appears that 959, 960, and 961 are all lost in D. virilis. For 961, the reported mojavensis and grimshawi orthologs are confident although neither of these orthologs has any small RNA reads.
11 virilis reads mapped onto the 961/dwil ortholog. It would still be good to validate that this miRNA is lost in the virilis group. |
| dme-mir-968 |
droWil1 |
Not found |
|
Yes |
Sequence the 1002 and 968 virilis orthologs |
This miRNA is located antisense to an intron, however, I could not locate the gene or intron that emcompassed both this miRNA nor 1002, an adjacent miRNA.
Searching for the correct droVir3 ortholog with the droWil1 ortholog was unsuccessful. It should be noted that 968/dmoj is also missing yet d68/dgri looks confident and has one 15 reads.
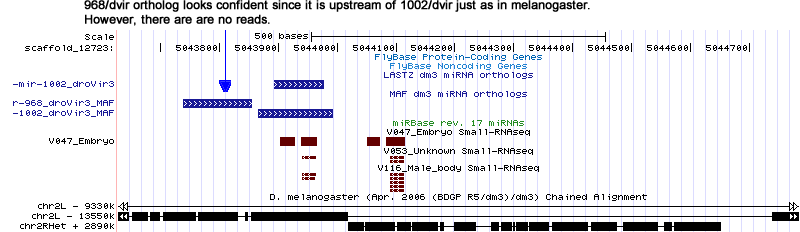
Many virilis reads mapped to the grimshawi ortholog. For example the read V047_15_28649 corresponds to the mature sequence "TAAGTAGTAACCATTAAGAGGTTG". |
| dme-mir-1002 |
droWil1,droGri2 |
Not found |
|
Yes |
Sequence the 1002 and 968 virilis orthologs |
Many virilis reads mapped to the willistoni ortholog. For example, the reads V047_109_2049 corresponds to the mature sequence "TTAAGTAGTTAATACAAGGGCGA". It appears that the star sequence may be V041_168952_1
which corresponds to "GCATTGTGTGAGCTACTTC" |
| dme-mir-978 |
droWil1, droGri2 |
Found |
|
N |
NA |
>gnl|ti|376844870:534-649
A------------------------CAGGCACAGCCGTACTCTACGCTTTTGGGAACGAGCTCTTTGACGCACTCGGTTCCATTGCCGTTGAGTAGAGCTGTCCTGATGCATATCTCAACGT------ATTGATTCAAACAAGAA
>dme-mir-978_droGri2
AAGTACGGCAGTTCGATAACCAACACAGGTCCAGCCGTATTCAGCGCTTTTGGAAGTAAGGAACATGATTCTCCTAGTTTCAATGCCGCTGAGTAAAGCTGTCCCAATGCAAATCGCAACGTAACTGAATCGAATCAATCAATAA
The correct ortholog was found by searching the virilis trace reads with the grimshawi ortholog. A simlar search with the willistoni ortholog did not uncover any hits. Note that the mojavensis ortholog was not found. |
miRNA Name |
Query Species |
Result |
Fragment for Cloning |
Experiment Needed |
Experiment decision |
Sequence/Notes |
| dme-mir-312 |
droGri2 |
Found |
scaffold_6496:9090108-9091422
(1314nt) |
Y |
Cluster will be resequenced. |
Searching with the grimshawi ortholog against the mojavensis genome trace reads found the sequence that is supposed to be 313/moj.
>dme-mir-312-droGri2
AAGCAAATCTTCAGTTTTTCGGCTGTGGTTAGTGTCAATTCTTTTTATTTGACAAGCATTGCACTCTTCACGGCCGGATAAATGAGGGTCT
>gnl|ti|490408270:305-377
-------------GTTTTCCGGTTGTGAATTGTGTCAATTCTTTTTATTTTACAAGTATTGCACTTTTCACGGCCGGAAAAAGGA------
This observation is actally also true for persimilis where the 313/dper ortholog was found when searching for the 312/dper ortholog with 312/dp4 query. |
| dme-mir-263b |
droVir3, droGri2 |
Not found |
Genomic coordinate is unknown. We may have to design primers using the virilis or grimshawi orthologs. |
Y |
Will be resequenced. |
Searching with the droVir3 or droGri2 ortholog in the droMoj3 trace reads did not turn up any hits.
No good chains exist for this miRNA in the droMoj3 assembly. This miRNA lies in an intron of the gene dme/CG32150. Flybase does not report an ortholog for this gene in mojavensis, however does report orthologs in virilus (dvir\GJ11223) and grimshawi (dgri\GH14363).
I tried looking for flanking genes to dme/CG32150 and found two with flybase-reported orthologs in D. mojavensis- dmoj/GI13967 and dmoj/GI11303. Next I looked for gaps in the intermediate region of these two orthologs and found 5. The genomic coordinates for the top 3 are shown in the image below.
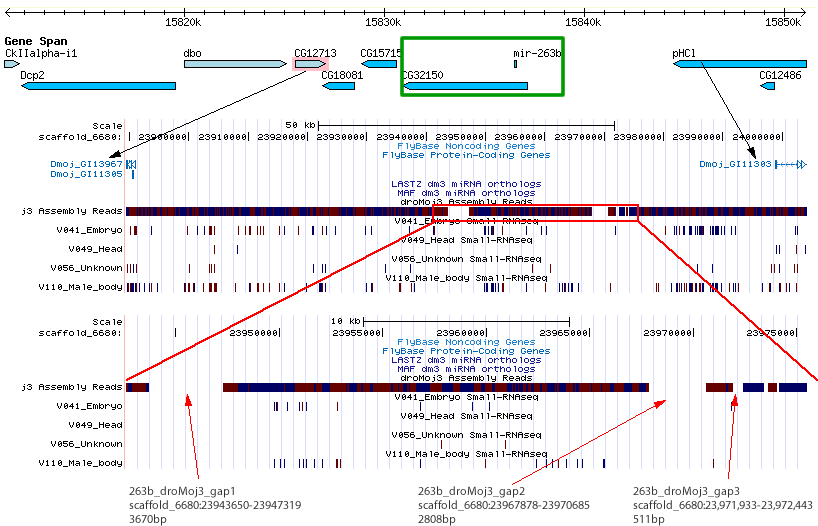
We should seqeuence any of these regions.
Do the mojavensis reads map to to the 263b/dvir ortholog?
Yes. Many reads from the mojavensis library maps to the virilus ortholog. The most abundant uniquely mapped reads are V041_122_1442, V110_640_2972, and V049_149_2107. The reads all have the same 23nt DNA sequence "CTTGGCACTGGGAGAATTCACAG". Note that these reads did not map to the droMoj3 genome assembly. Even more surprising, the DNA sequence of these reads could not be found via LASTZ in the droMoj3 trace reads either. |
| dme-mir-968 |
droWil1 |
Not found |
Actual genomic location is unknown. We may have to design primers using the willistoni ortholog. |
Y |
Nothing done |
The MAF reports the 968/dmoj ortholog in the correct position. That is, upstream of the 1002 ortholog just as in every other species. However this ortholog has no reads:
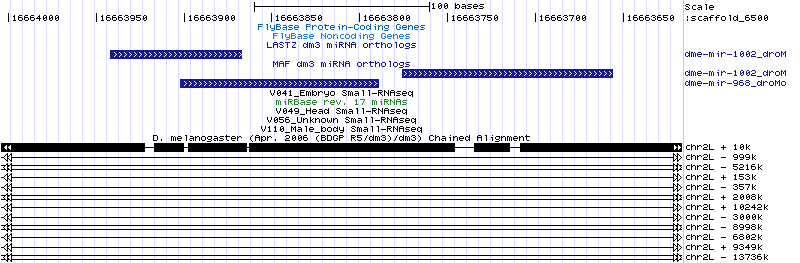
The mojavensis read V041_35_10366 mapped to the willistoni ortholog with 2 mismatches. This read is "TAAGTAGTAACCATTAAAAGGTCG" and corresponds to the mature miRNA for the other species. This is a good indication that the correct ortholog remains to be found. Searching for the correct droVir3 ortholog with the droWil1 ortholog was unsuccessful. It should be noted that 968/dvir is also missing yet 968/dgri looks confident and has 2 perfectly-mapped reads. |
| dme-mir-1002 |
droWil1, droGri2 |
Not found |
|
Y |
Nothing done |
The mojavensis reads V041_58_4018 mapped to the willistoni ortholog and corresponds to the mature sequence "TTAAGTAGTTGATACAAAGGCGA" with 2 mismatches. |
| dme-mir-978 |
droWil1, droGri2 |
Not found |
scaffold_6328:2896355-2898115
(1760nt) |
Y |
Nothing done |
The correct mojavensis ortholog could not be found by searching the trace data with the willistoni or grimshawi orthologs. However, the virilis ortholog was found. Given that the grimshawi and virilis orthologs are confident, it is unlikely that the mojavensis ortholog is genuinely lost. Resequencing is needed for this miRNA.
There are also many unannotated miRNAs in the vicinity of 978/dmoj. Could one of these be 978/dmoj?
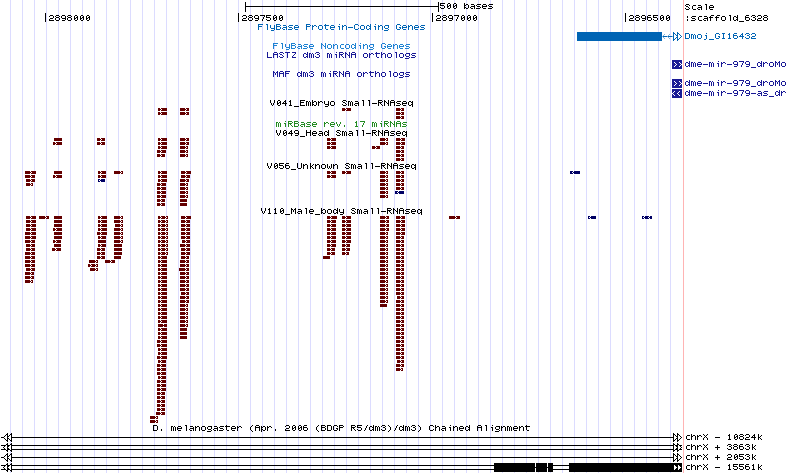
|