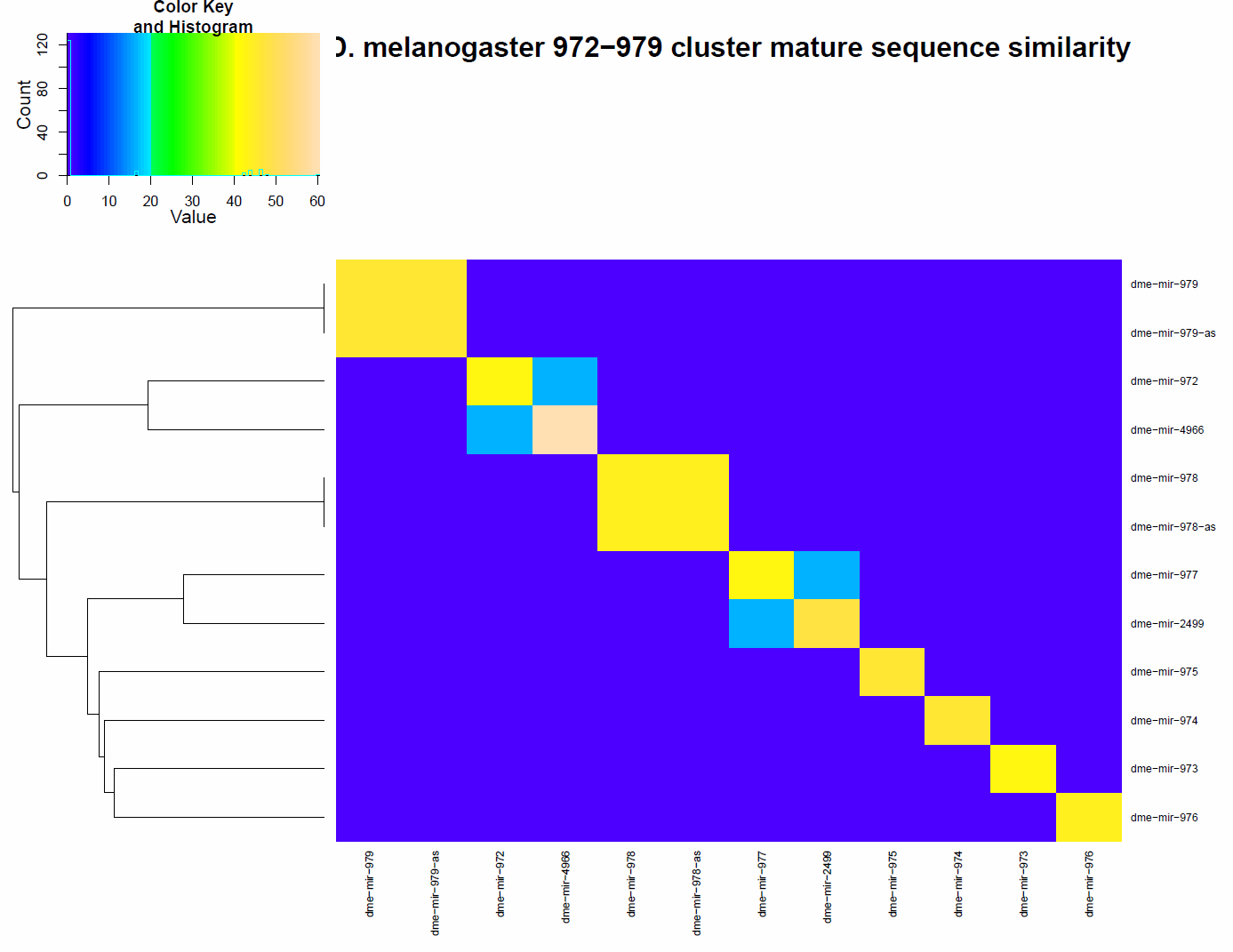
The heatplot below shows the similarity of the mature region of each miRNA within this cluster. Similarity was computed from BLASTN. It is identical to the bitscore reported from BLAST. As shown in this figure, all the miRs (i.e. mature regions of the miRNA) within this cluster are distinct despite several of the miRNA being adjacent to each other on the X chromosome.

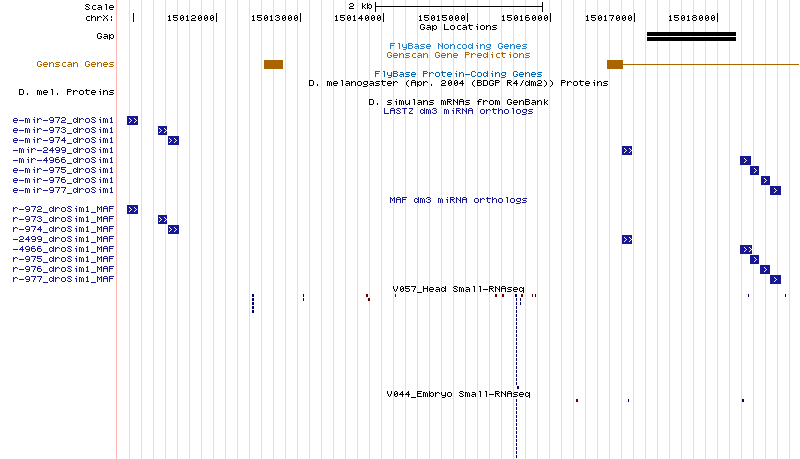
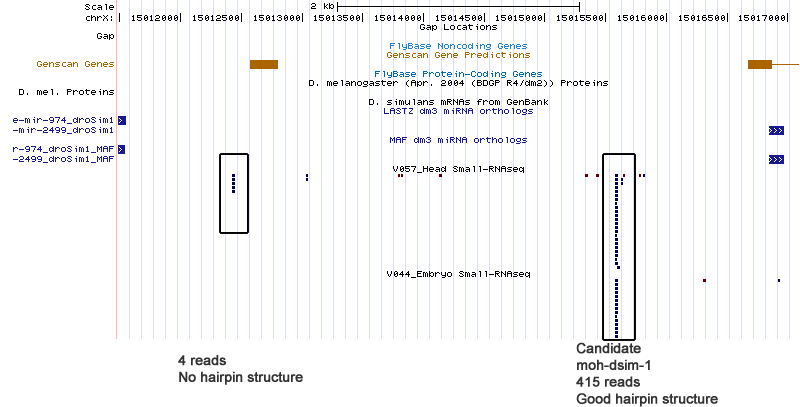
Below, you will find a series of UCSC browser screenshots for each of the candidate miRNA discovered using the small RNA-seq data. The list is broken down by species.
Candidates
| Name | Coordinate |
|---|---|
| moh-dsim-1 | chrX:15015576-15015647 |
Cluster view
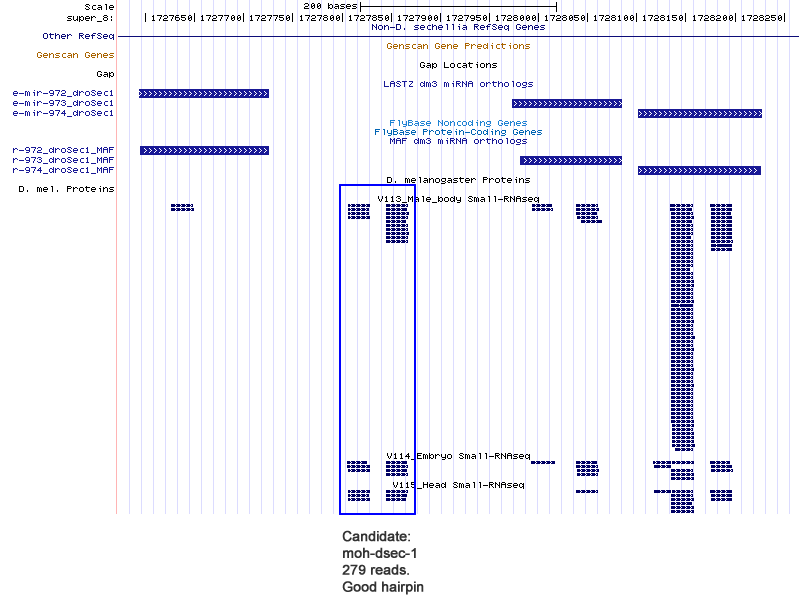
Candidates
| Name | Coordinate |
|---|---|
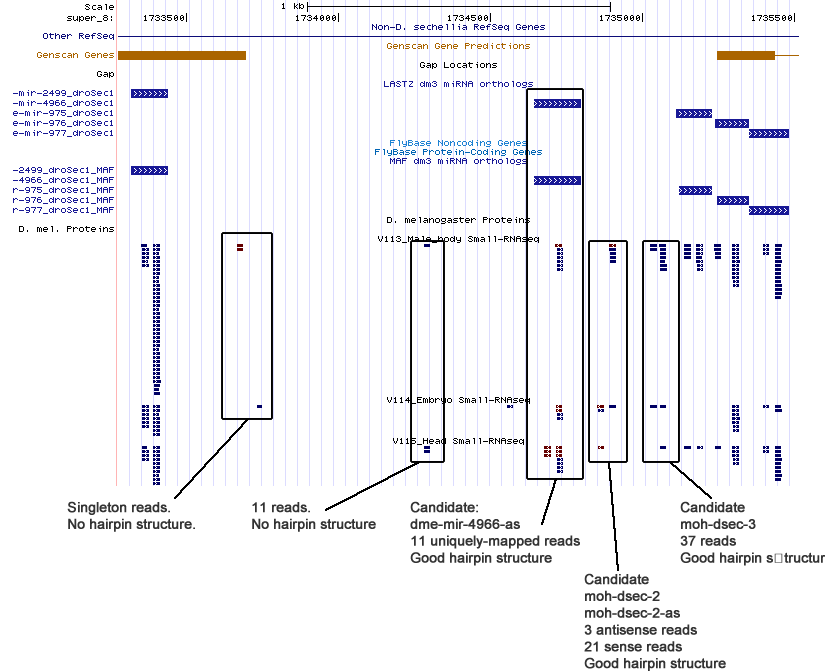
| moh-dsec-1 | super_8:1727806-1727868 + |
| dme-mir-4966-as | super_8:1734676-17347376 - |
| moh-dsec-2 | super_8:1734853-1734914 + |
| moh-dsec-2-as | super_8:1734852-1734911 - |
| moh-dsec-3 | super_8:1735027-1735079 + |
Cluster view


Cluster view
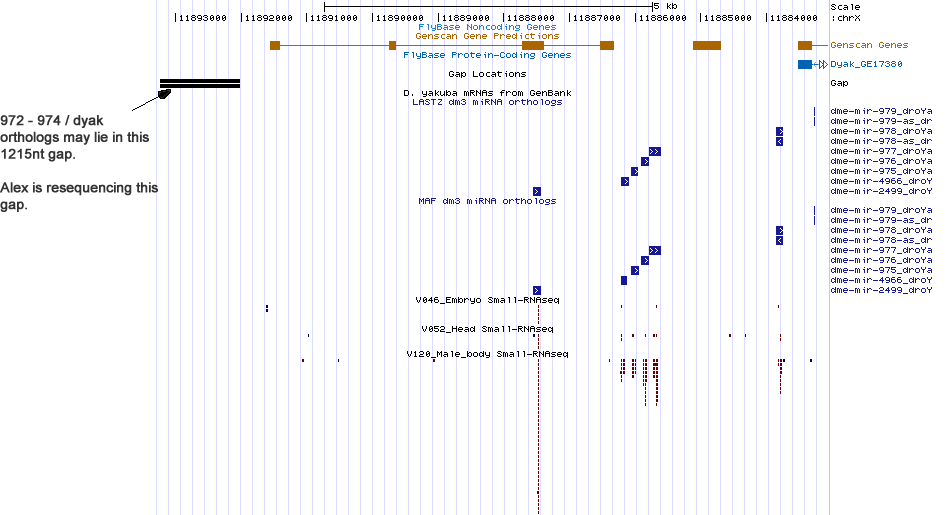
No new miRNA discovered in this cluster.
Candidates
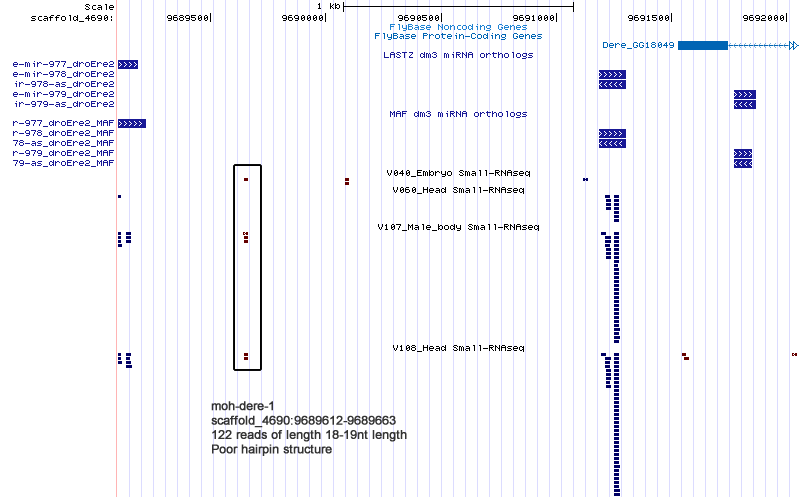
| Name | Coordinate |
|---|---|
| moh-dere-1 | scaffold_4690:9689612-9689663 |
Cluster view



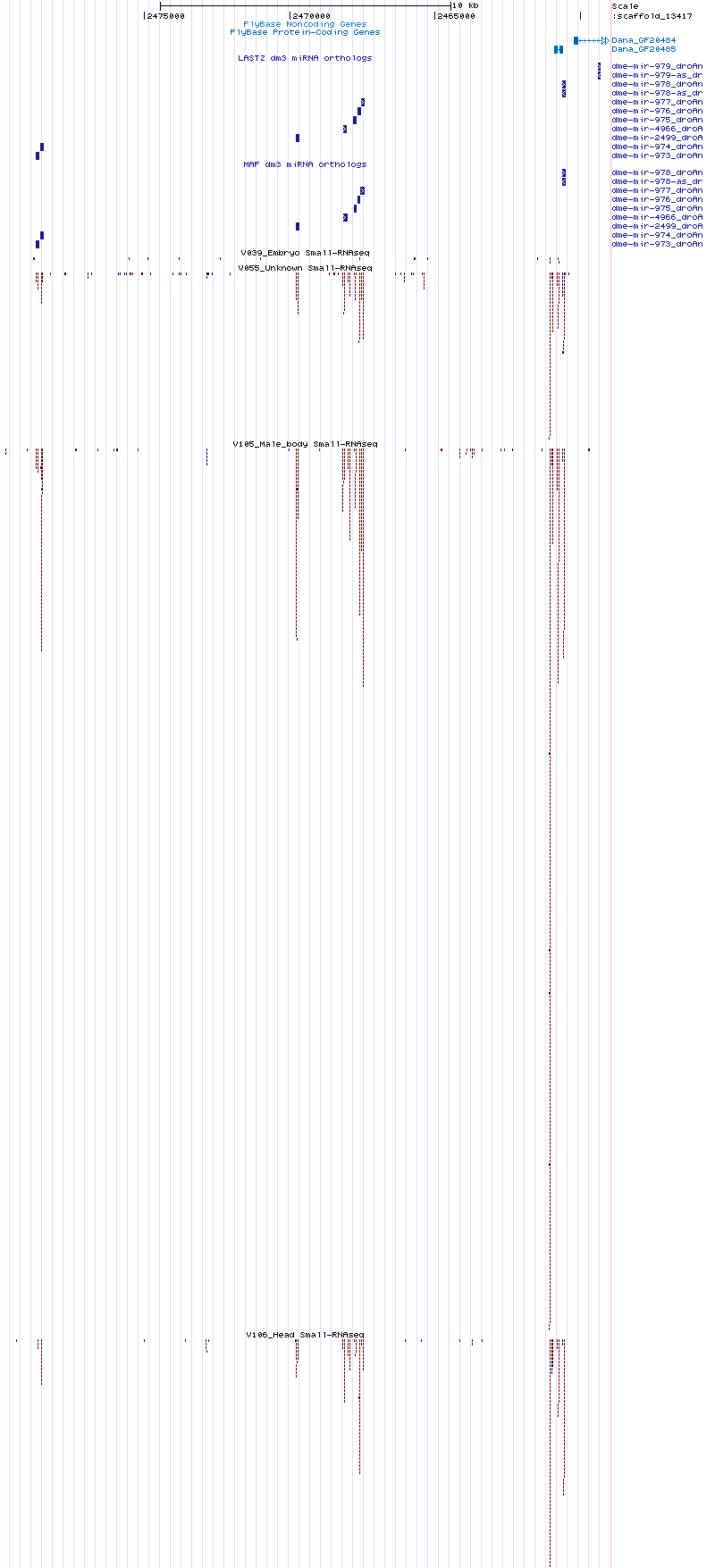
Candidates
| Name | Coordinate |
|---|---|
| moh-dana-1 | scaffold_13417:2472648-2472875 |
| moh-dana-1as | scaffold_13417:2472784-2472893 |
| moh-dana-2 | scaffold_13417:2467915-2468008 |
| moh-dana-3 | scaffold_13417:2461028-2461095 |
| moh-dana-4 | scaffold_13417:2460923-2460984 |
| moh-dana-5 | scaffold_13417:2460722-2460792 |
Cluster view
scaffold_13417:2458998-2479950
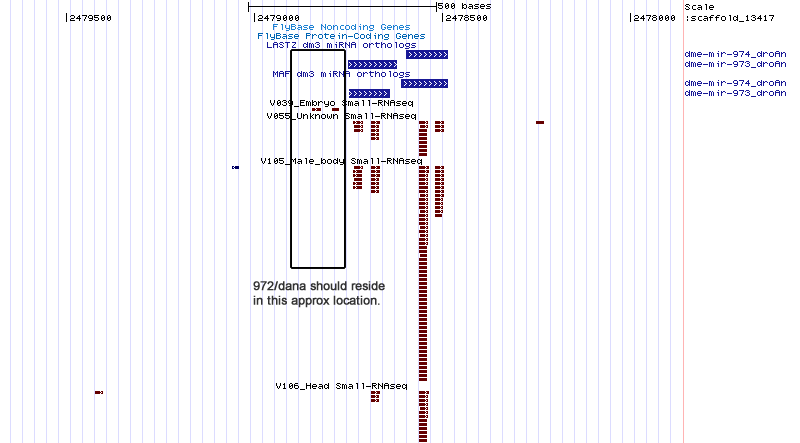
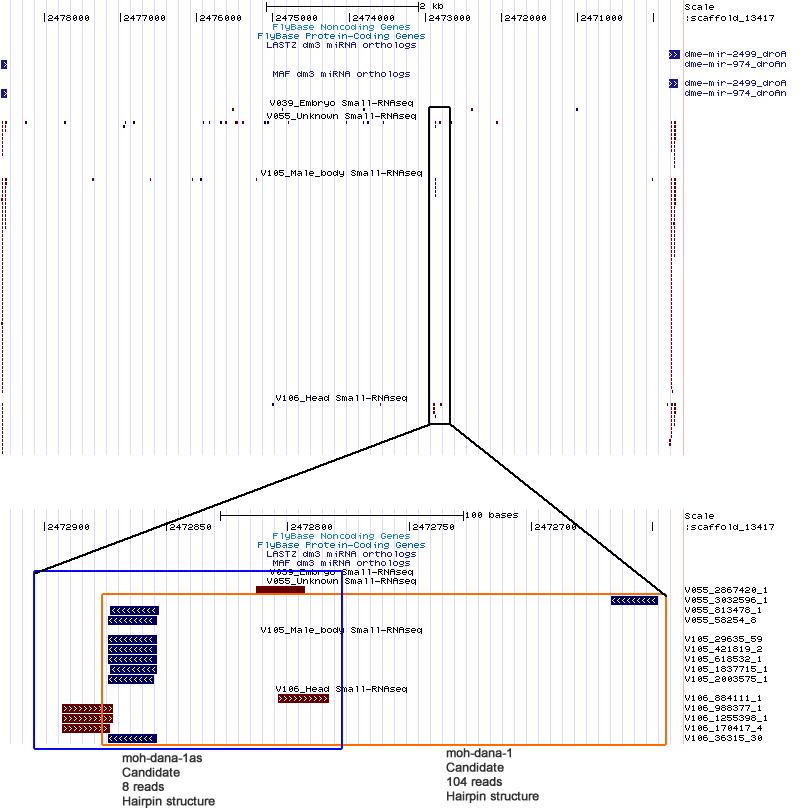

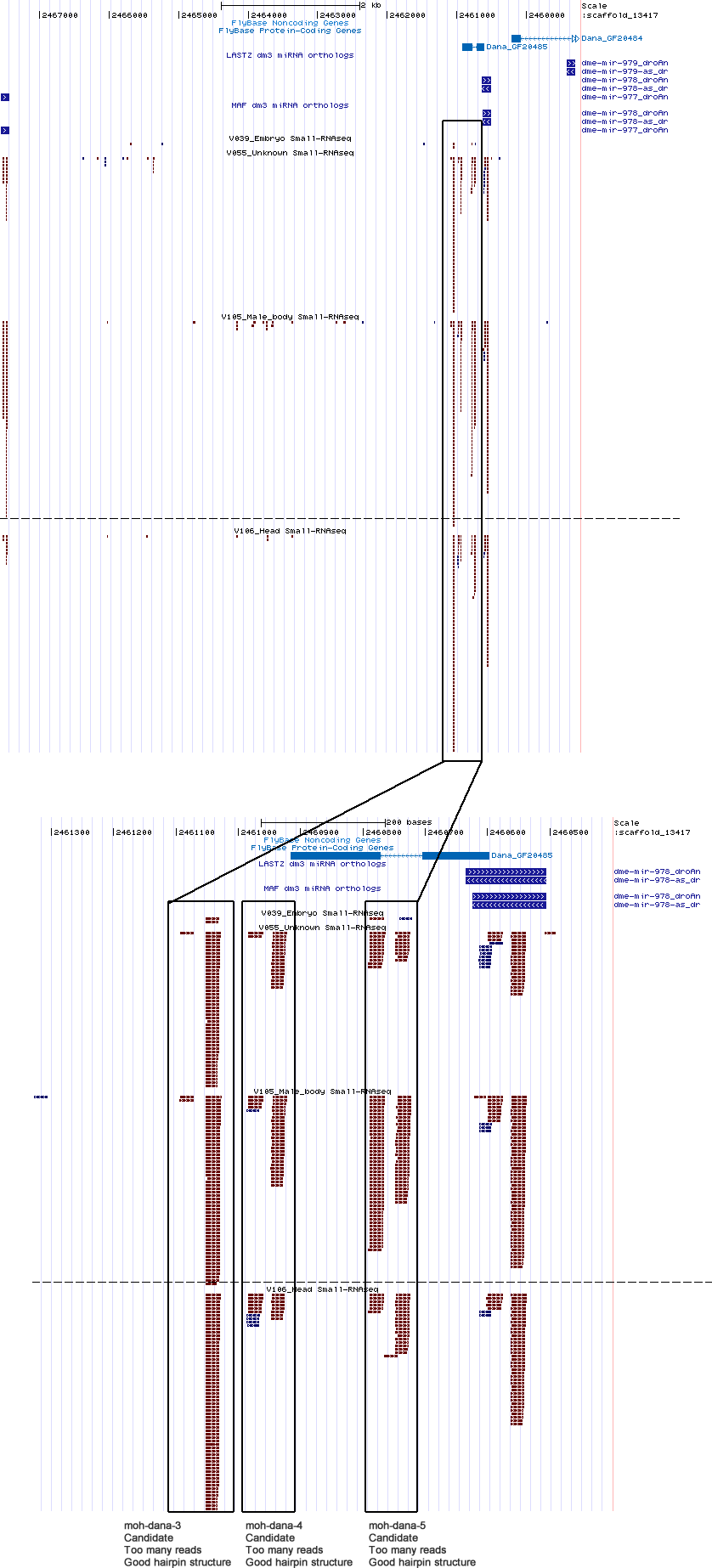
No orthologs found.
No orthologs found.
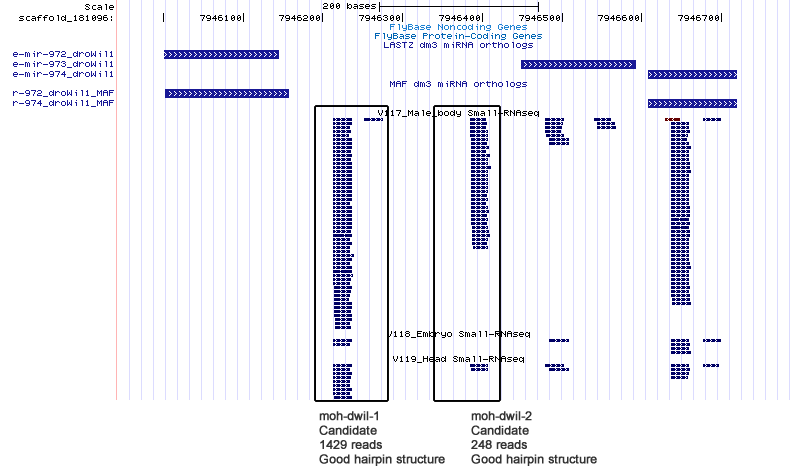
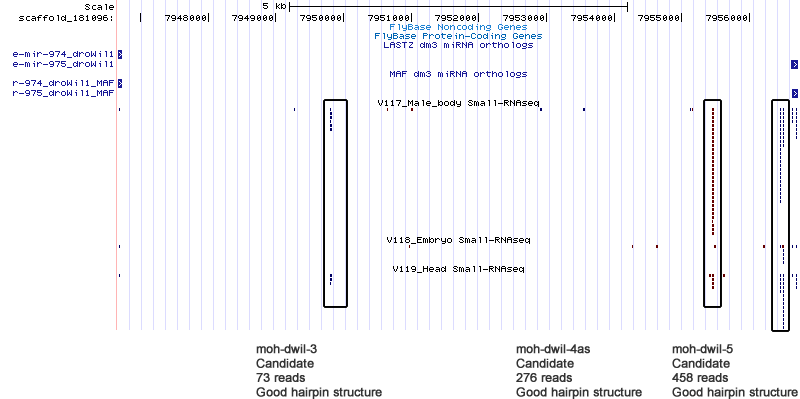
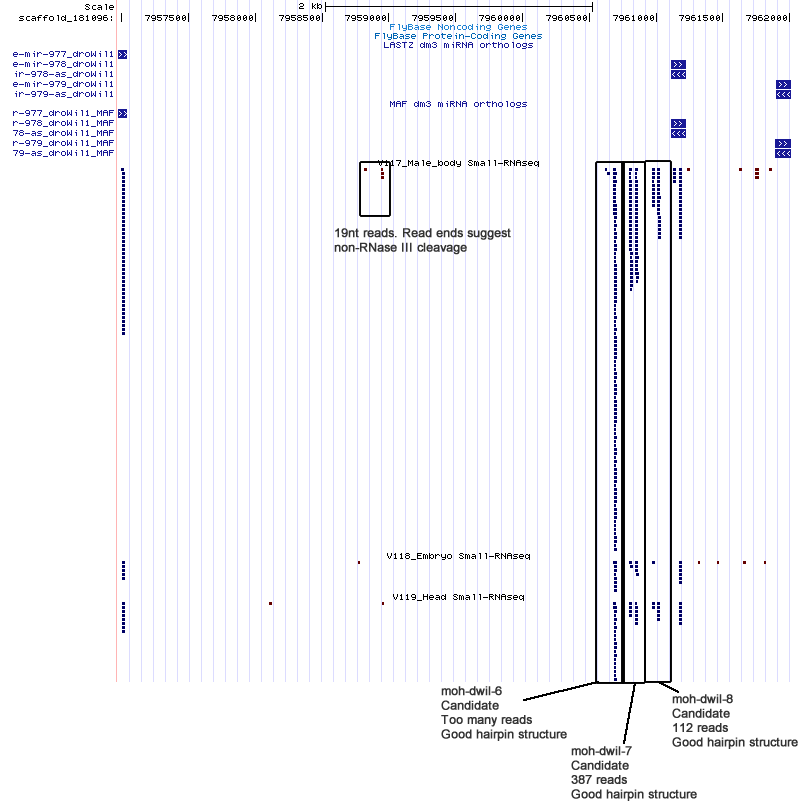
Candidates
| Name | Coordinate |
| moh-dwil-1 | scaffold_181096:7946212-7946276 |
| moh-dwil-2 | scaffold_181096:7946382-7946453 |
| moh-dwil-3 | scaffold_181096:7949753-7949826 |
| moh-dwil-4 | scaffold_181096:7955395-7955470 |
| moh-dwil-5 | scaffold_181096:7956441-7956514 |
| moh-dwil-6 | scaffold_181096:7960633-7960716 |
| moh-dwil-7 | scaffold_181096:7960793-7960871 |
| moh-dwil-8 | scaffold_181096:7960964-7961040 |
| dme-mir-973-droWil1 | scaffold_181096:7946479-7946569 |
| dme-mir-974-droWil1 | scaffold_181096:7946634-7946703 |
| dme-mir-975-droWil1 | scaffold_181096:7956619-7956705 |
| dme-mir-976-droWil1 | scaffold_181096:7956783-7956870 |
| dme-mir-977-droWil1 | scaffold_181096:7956951-7957029 |
| dme-mir-978-droWil1 | scaffold_181096:7961127-7961200 |
Cluster view
scaffold_181096:7945397-7962301
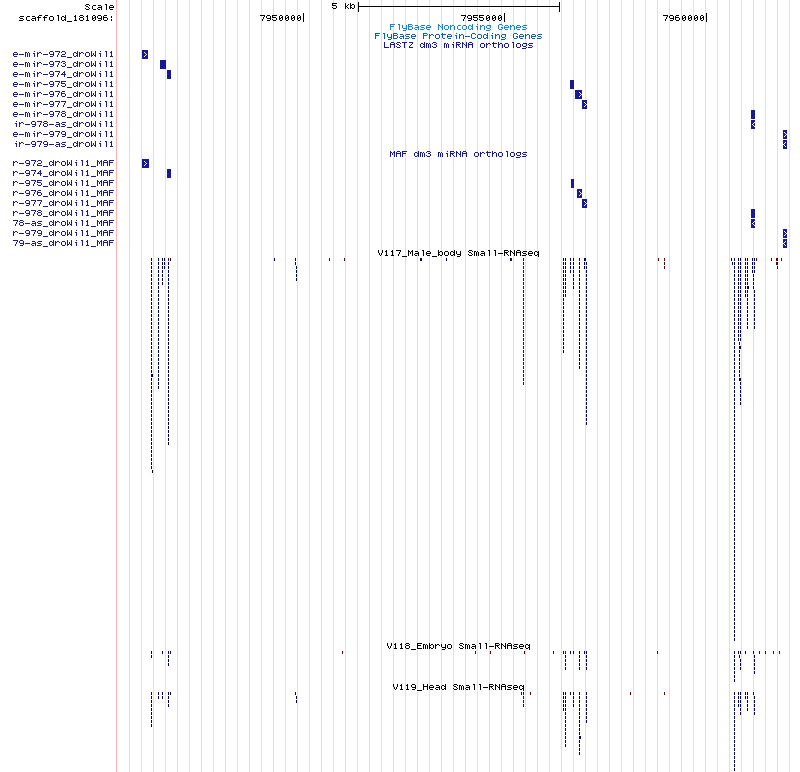
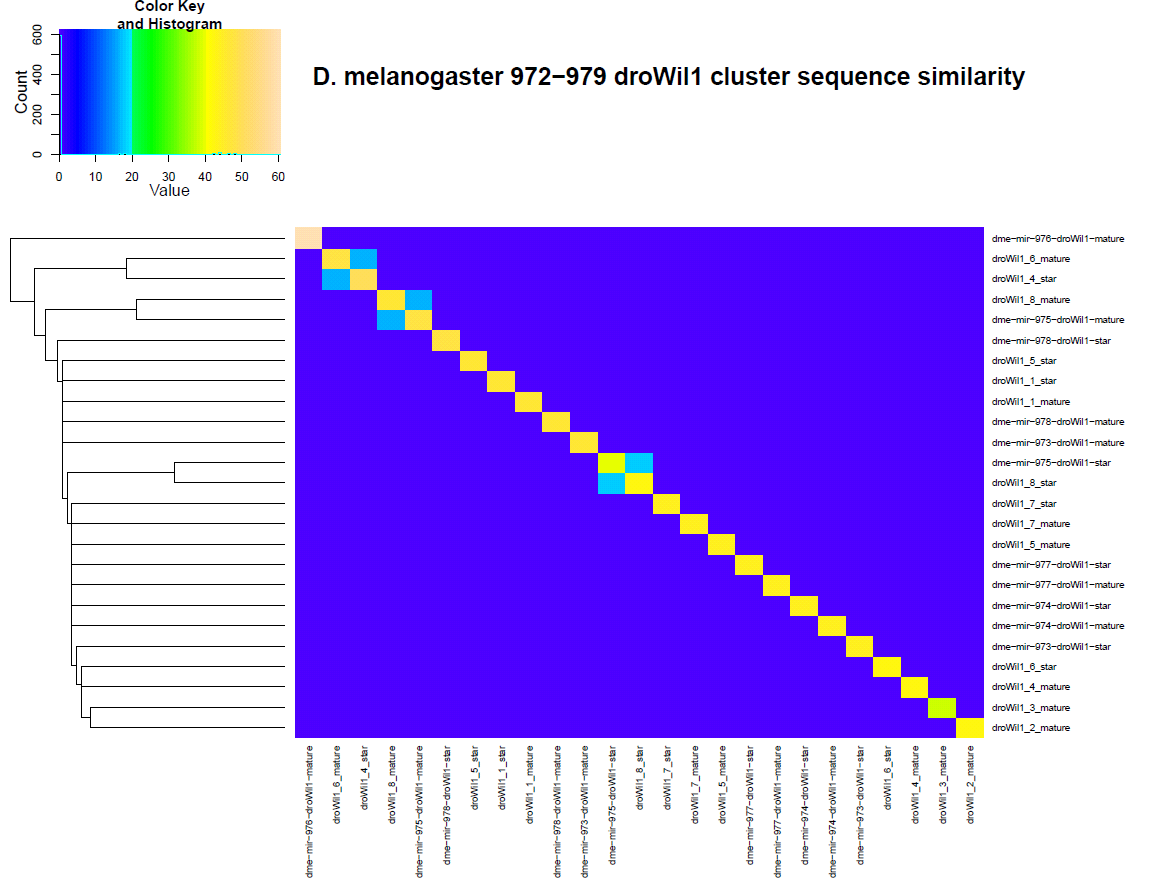
The similarity comparison between these new candidates and the known orthologs can be seen in the below heatmap. What becomes apparent immediately is the similarity between the dme-mir-975-droWil1 ortholog and that of the candidate locus 8.
The similarity of these two loci becomes clearer in their alignment:
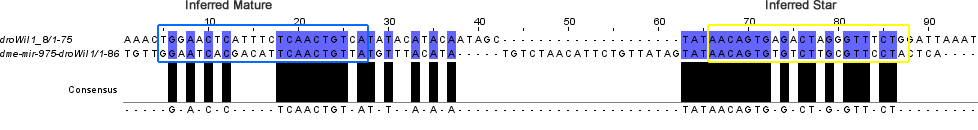
Candidates
| Name | Coordinate |
| droVir3_1 | scaffold_13042:917881-917949 |
| droVir3_2 | scaffold_13042:917745-917815 |
| droVir3_3 | scaffold_13042:912568-912655 |
| droVir3_4 | scaffold_13042:912399-912465 |
| droVir3_5 | scaffold_13042:907097-907164 |
| droVir3_6 | scaffold_13042:903357-903448 |
| droVir3_7 & 7as | scaffold_13042:903190-903272 |
| droVir3_8 | scaffold_13042:903013-903106 |
| droVir3_9 | scaffold_13042:902868-902935 |
| droVir3_10 | scaffold_13042:902694-902766 |
| dme-mir-973-droVir3 | scaffold_13042:918354-918425 |
| dme-mir-974-droVir3 | scaffold_13042:917565-917638 |
| dme-mir-975-droVir3 | scaffold_13042:906925-906995 |
| dme-mir-976-droVir3 | scaffold_13042:906747-906812 |
| dme-mir-977-droVir3 | scaffold_13042:906580-906653 |
Cluster view
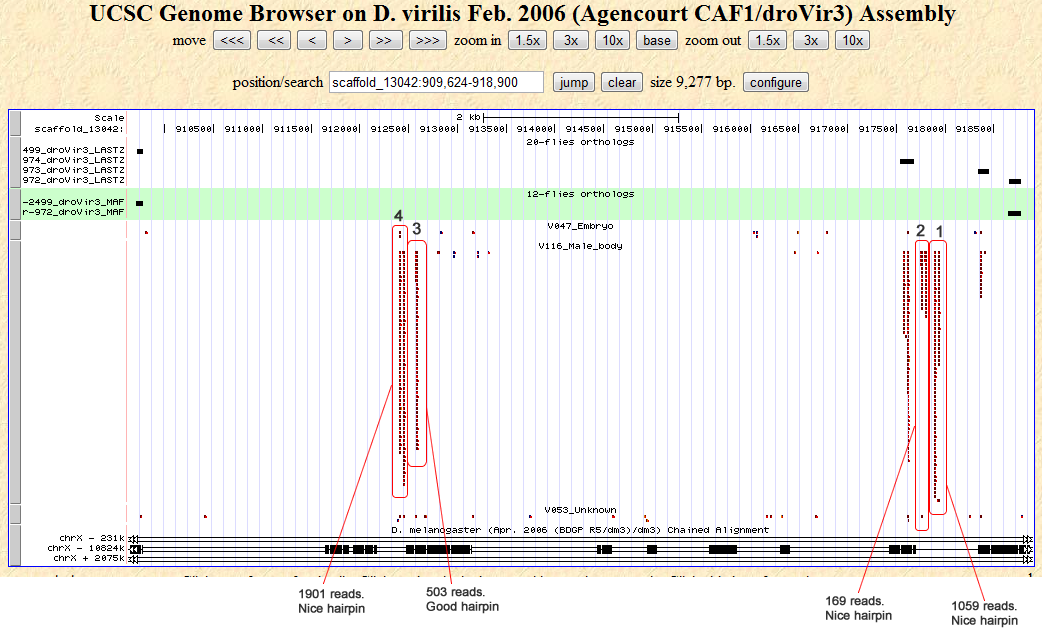

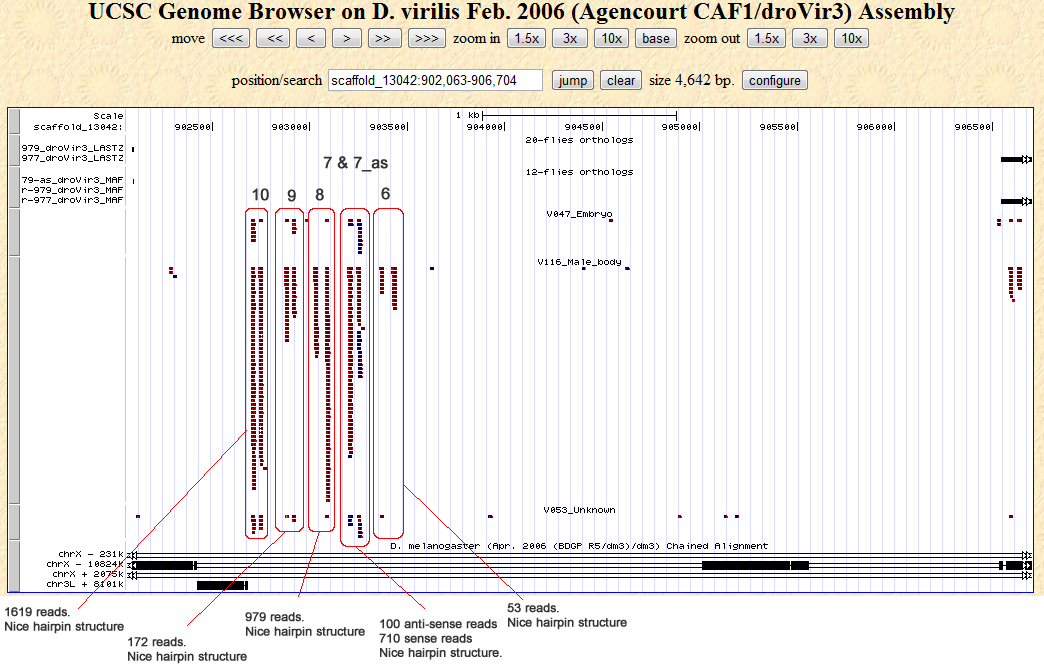
Notes:
Firstly, the droVir3_3 candidate locus may be a duplicate droVir3_4, since all the reads at the position scaffold_13042:912568-912655 are repeats and also map to droVir3_3. An example of two such high read count sequences are:
| Read ID | V116_22290_103 |
| Read Sequence | GGATAGCTACCGTTCAATGTACAA |
| First hit | scaffold_13042:912574 (droVir3_3) |
| Second hit | scaffold_13042:912441 (droVir3_4) |
| Read ID | V116_13609_197 |
| Read Sequence | TAGCTACCGTTCAATGTACAA |
| First hit | scaffold_13042:912577 |
| Second hit | scaffold_13042:912444 |
Secondly, the similarity between the candidate loci and the virilus orthologs can be seen in the heatmap below. I should point out that he similarity between dme-mir-974-droVir3 ortholog and droVir3_2 candidate locus.
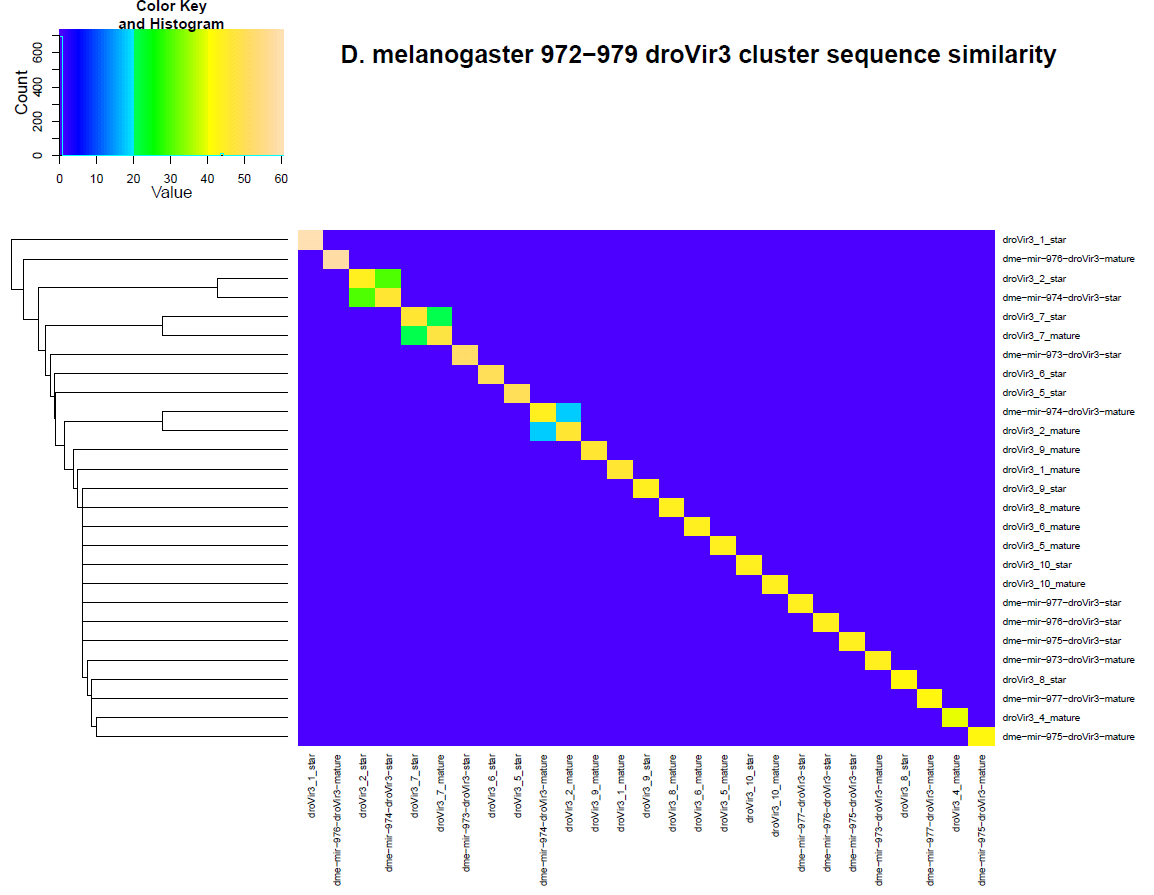
Here is the alignment of candidate locus droVir3_2 with dme-mir-974-droVir3 ortholog:
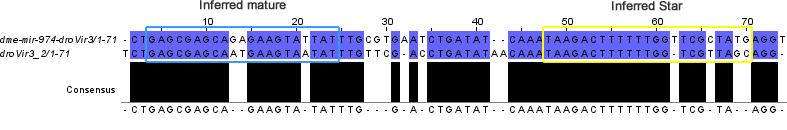
Thirdly, I'm still puzzled why the droVir_7 candidate locus' mature and star sequences are so similar. What is even more bizarre is that there are both sense and antisense reads for this locus. How did this locus evolve? What is its expression vehicle given that all miRNAs in this location are on the negative strand?
Candidates
Cluster view
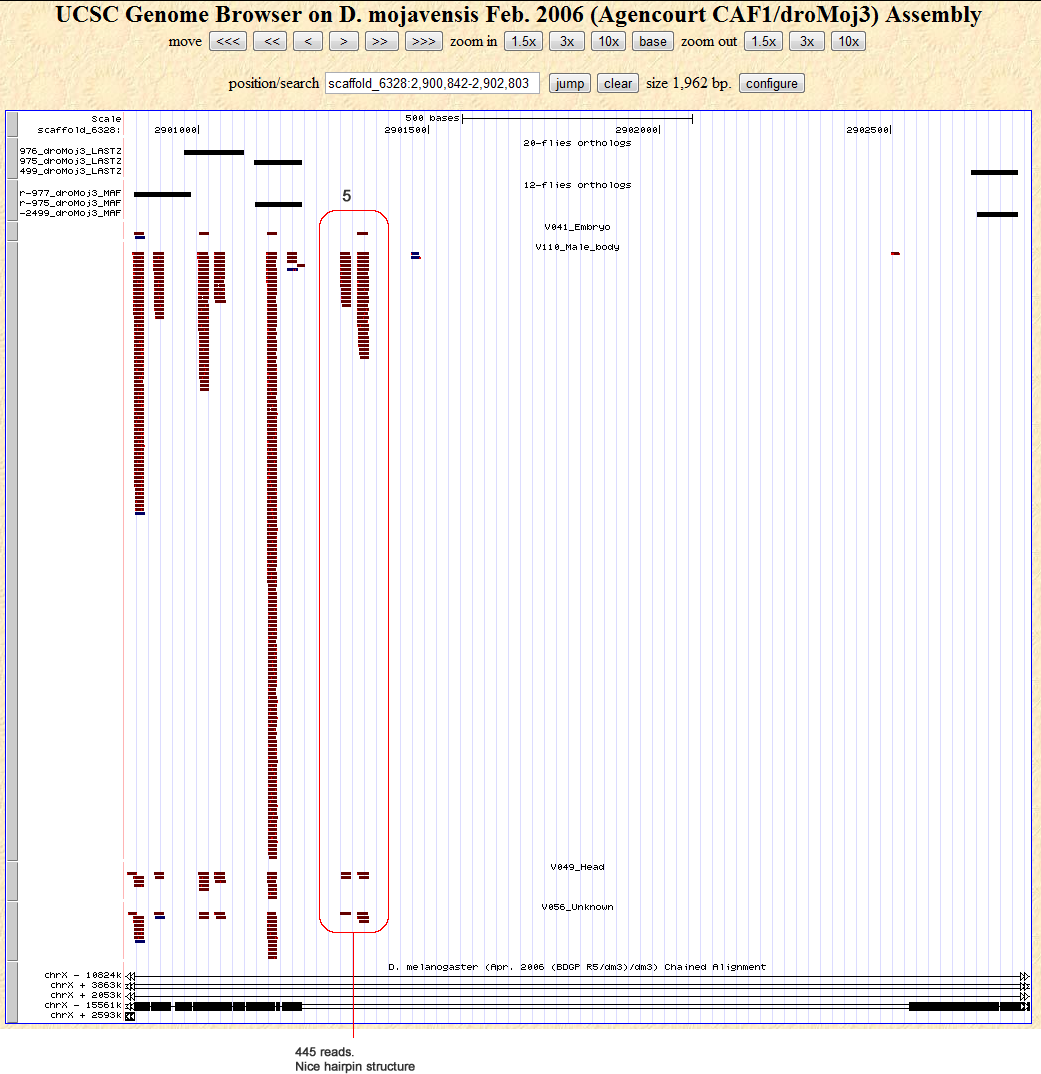
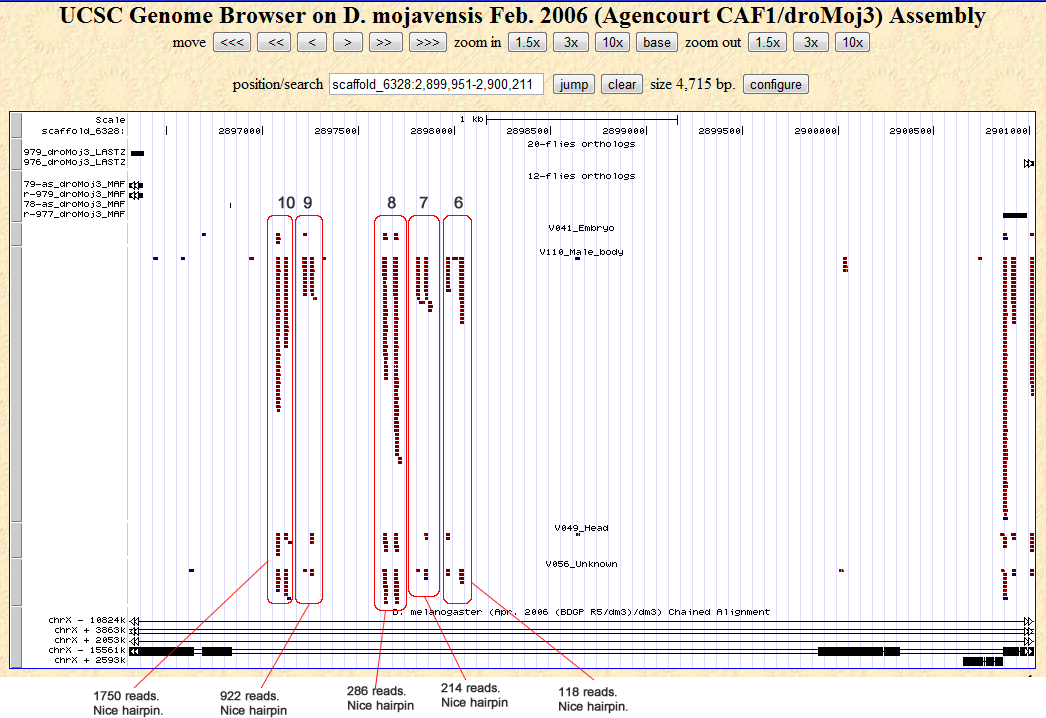
Candidates
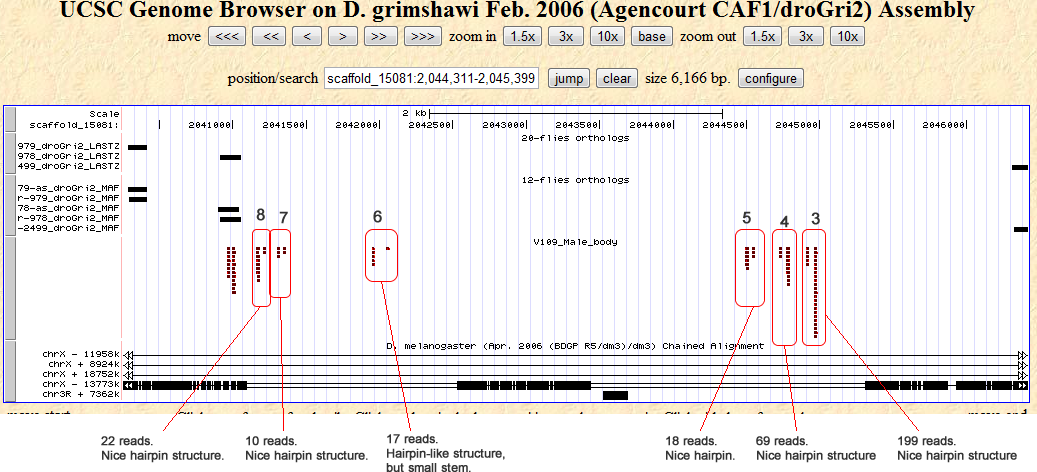
Cluster view
Updated: 12/12/2011 by Jaaved