In the table below, I've summarized the list of variants found genome-wide and those that overlap with D. melanogaster orthologs. Note that there are 342 total known and candidate miRNAs in D. melanogaster.
| Species | Total Variants detected | Variants in Both | Variants in sRNAseq | Variants in Trace | Variants overlap with dm3 miRNA orthologs | Variants in Both | Variants in sRNAseq | Variants in Trace | Valid Variants | Number of orthologs | Number of orthologs with overlaps | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 | 678685
[link] |
52 |
286 |
678347 |
133 [link] |
2 | 4 | 127 | 133 | 321 |
78 |
Simulans is a mosaic genome. Therefore, many (127/133) variants detected from only trace data. |
| droSec1 | 44259 [link] |
19 |
10264 |
33976 |
4 [link] |
0 | 4 | 0 | 4 | 339 |
3 |
|
| droYak2 | 27708
[link] |
13 |
1298 |
26397 |
1 [link] |
0 | 1 | 0 | 1 | 328 |
1 |
|
| droEre2 | 41225 [link] |
10 |
6348 |
34867 |
16 [link] |
1 | 5 | 10 | 14 | 332 |
8 |
|
| droAna3 | 91346 [link] |
40 |
11869 |
79437 |
3 [link] |
0 | 3 | 0 | 3 | 292 |
2 |
|
| dp4 | 106803 [link] |
83 |
8904 |
97816 |
8 [link] |
0 | 6 | 2 | 7 | 276 |
7 |
|
| droPer1 | 40702 [link] |
2 |
4470 |
36230 |
4 [link] |
0 | 4 | 0 | 3 | 266 |
3 |
|
| droWil1 | 76942 [link] |
262 |
7620 |
69060 |
4 [link] |
0 | 4 | 0 | 0 | 249 |
4 |
All variants found appear to be RNAediting. |
| droVir3 | 104930 [link] |
19 |
9525 |
95386 |
5 [link] |
0 | 4 | 1 | 5 | 247 |
4 |
|
| droMoj3 | 80342 [link] |
43 |
13153 |
67146 |
5 [link] |
0 | 4 | 1 | 5 | 245 |
5 |
|
| droGri2 | 286111 [link] |
57 |
3090 |
282964 |
28 [link] |
0 | 2 | 26 | 28 | 240 |
20 |
Similar to droSim1, many (26/28) variants detected from the trace data. |
Orange column- Variants found genome-wide. This list is further broken down by variants found in both RNAseq and trace reads and in each individually.
Blue column- Variants that overlapped with miRNA ortholog coordinates. This list is also broken down by variants found in both RNAseq and trace reads and in each individually.
Pink column- Variants in the blue column that were validated by visual inspection to be valid
Purple column- Number of miRNA orthologs containing a variant.
Variant- A disagreement between the reference base and the most likely base supported from the sRNAseq data and the assemblies' trace data. A variant is called if less than 50% of the sRNAseq reads or trace reads support the reference base. As of yet, INDELS are not accounted for in this definition. However, INDELS are sparse throughout the small RNAseq reads and I don't believe there is a significant total amount.
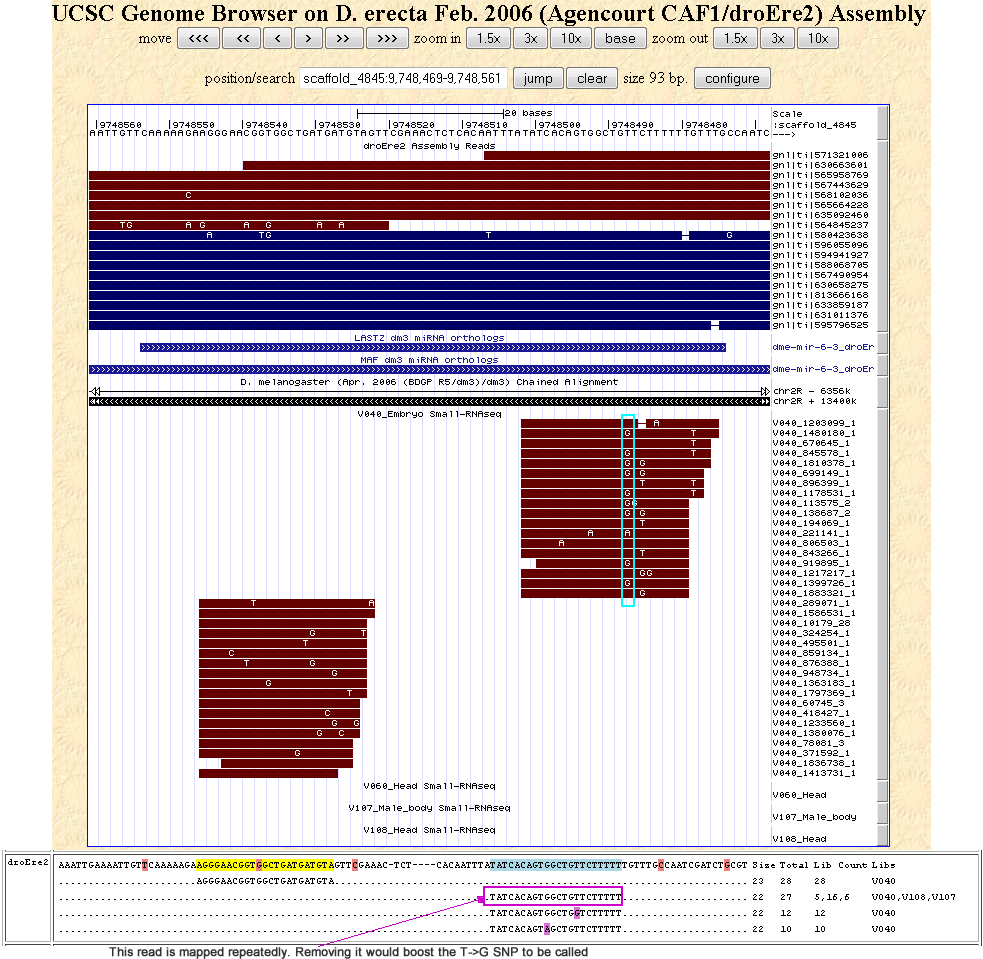
If you'd like to take a look at the pileup of reads per genomic position, I've uploaded the trace and sRNAseq reads onto my personal genomic browser at Jaaved's UCSC Genome Browser. Tracks for the D. melanogaster miRNA orthologs are also visible per genome.
Without mitigating errors in the pileup and variant detection analysis, hundreds of variants per species would be found. I've found that many of these cases appear to be false-positive hits for two main reason.
Many of the short reads were found to crossmap to various regions of the genome especially to areas with miRNA families. Instead of focusing on all mapped reads, I instead looked at the more confidently mapped ones. BWA reports the confidence of each read mapping (called the MAPQ score), which can be interpreted as a Phred-like score. I chose a confidence score of at least 1. As an explanation of what this score means, it is the probability that a mapping is wrong, and the score equals [ 10^(score / -1) ]. For reference, a MAPQ score of 0 indicates that P(wrong) = 1, a MAPQ score of 1 indicates P(wrong)=0.79, and score of 37 means P(wrong) = 0.0002. As you can see, reads with score of 0 should not be trusted.
There is also a fine balance between deciding when to use all reads and when use to only uniquely mapped reads. The case of artificially inferring supurious variants by using all mapped reads has already been stated. Vice verser, using uniquely mapped reads could also convolute the variants called. For example, for miRNAs with identical mature or star sequences, such as 6-1, 6-2 and 6-3, removing repeated reads may reduce the total read coveverage at a particular position, thus boosting the fraction of mismatch bases from suboptimal reads. In 6-3 below, we see from the browser screenshot that a variant is A->C SNP can be inferred, but we see from the pileup of all reads, that this variant is not supported. But these similar miRNA cases are few and cand be dealth with on a case-by-case basis, so we'll use only uniquely mapped reads.

In addition to cross-mapped reads, the orthologs for some miRNAs (especially those belonging to the same family) appear to overlap. This will convolute the estimates of miRNAs with variants.. Here is an example, where the D. sechelia ortholog of dme-mir-981-1 overlaps with several other orthologs:

Orthologs reported as the best hit by LASTZ may actually overlap with the same ortholog found for another miRNA belonging to the same family. As a tie breaker, I've looked in depth at overlapped orthologs and either marked one of the overlapping orthologs as missing (which is more common) or looked at the lower confident LASTZ hits and chosen the ortholog which would fit in the approximate correct genomic locale relative to the other neighboring orthologs.
Finally, many of the false-positive hits of the variant detection pipeline turned out to be 3' untemplated additions, or RNA editing events. For example, the D. persimilis ortholog for dme-mir-1010, a mirtron. Notice that the C at position 8195080 is supported by the 3 assembly reads, but not by any of the RNAseq reads mainly due to 3' uridylation.
We would like to differentiate these RNA editing events from the bona fide genomic variants for this analysis. Therefore, any variant detected on the end, or one base pair away from the ends of reads were removed.
Updated: 8/15/2011